
Review: biological implications of oncogenic rearrangements in non-small cell lung cancer
Introduction
Chromosomal rearrangements resulting in fusion proteins are a common oncogenic event in many human cancers. First discovered in hematological tumors, in the last three decades recurrent fusion oncoproteins have been found in solid tumors as well, using traditional methods such as cytogenetics, fluorescence in situ hybridization (FISH), and functional transforming assays. With the advent of next-generation sequencing (NGS) in the early 2000s, the number of recurrent gene fusions identified in solid tumors, including non-small cell lung cancer (NSCLC), has grown exponentially, and data emerging from The Cancer Genome Atlas (TCGA), the International Cancer Genome Consortium (ICGC) and individual studies have shown that gene fusions in solid tumors are more common than previously thought. To date, over 90% of gene fusions have been discovered in solid tumors by whole genome and transcriptome sequencing. However, only a fraction of them are “driver” events, with the majority being non-functional or insignificant bystanders. In fact, the presence of genomic fusions or rearrangements does not necessarily imply a functional protein product that can cause cancer development (1).
Lung cancer is the leading cause of cancer related mortality worldwide and constitutes an estimated 18.4% of all cancer deaths (2). Molecular characterization of lung cancer has been crucial for understanding its biology and is a key factor in improving diagnostics and developing effective treatments. NSCLC is the most frequent histological subtype, accounting for 85% of lung tumors. Among NSCLC, lung adenocarcinoma (LUAD) and squamous-cell carcinoma (LUSC) are the most common types (3,4).
Although several genetic alterations have been described in NSCLC, the oncogenic drivers of many lung cancers are still unknown. The most common genetic alterations thus far identified are mutations of the EGFR (17%) and KRAS (30%) genes. EGFR was the first targetable oncogene in NSCLC, and EGFR therapy has long served as the standard of care for lung cancer (5). KRAS, instead, was until recently a drug-orphan oncogene (6). In addition to mutations, recurrent oncogenic gene fusions are also common in NSCLC. These fusions occur mainly in the adenocarcinoma subtype, making up almost 10% of all LUADs, and frequently involve the Anaplastic lymphoma kinase (ALK) (5–6%), C-ros oncogene 1 (ROS1) (2%), Ret Proto-Oncogene (RET) (1%) and neurotrophic tyrosine kinase receptor (NTRK) (<1%) genes (5). Generally speaking, these genetic alterations are mutually exclusive, drive the initiation and progression of cancer, and are still present during sub-clonal diversification. This makes them an optimal target for therapy/treatment (7).
In lung cancer, both oncogenic fusions and EGFR mutations are found more frequently in patients with a light or non-smoking history. In addition, oncogenic fusions are mainly observed in young people, and occur more often in women than in men (6). Frequencies also vary by geographical region, with EGFR mutations and ALK fusions appearing more frequently in Asian than in Western populations. When considered individually, oncogenic fusions are relatively low-frequency events; collectively, however, they represent a substantial group of actionable driver oncogenes in LUAD (4). Their clinical importance has increased over the years because they define distinct subsets of NSCLC patients, and their identification has prompted the development of new diagnostic tests and several new drugs for the treatment of these patients (1,6,8).
This review presents an overview of the current state of knowledge about NSCLC characterized by oncogenic rearrangements and of the therapeutic options available for these tumors.
Oncogenic rearrangements in lung cancer
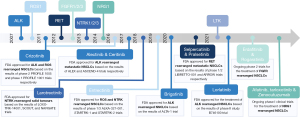
The first oncogenic fusion identified in NSCLC was ALK, in 2007. The tyrosine kinase inhibitor (TKI) crizotinib, originally developed to inhibit the c-MET tyrosine kinase receptor, showed remarkable activity against ALK-rearranged NSCLC in phase I–II clinical trials. Within four years, in 2011, crizotinib received FDA approval for the treatment of advanced ALK-rearranged NSCLC patients (9). Since then, second and third-generation ALK TKIs have also been developed, and treatment with TKIs has become the standard front-line therapy for advanced NSCLC (6,10). In addition, the encouraging results with crizotinib treatment for NSCLC provided a strong rationale for identifying other recurrent oncogenic fusions that could be targeted using already available TKIs. For example, when rearrangements of the ROS1 gene were found in NSCLC patients, the high similarity of the kinase domain of ROS1 and ALK proteins prompted several clinical trials with crizotinib that also showed some efficacy in ROS1 driven tumors (11). Subsequently, whole genome and transcriptome analyses revealed other oncogenic rearrangements including the RET, NTRK, neuregulin 1 (NRG1), fibroblast growth factor receptor (FGFR)1/2/3 genes—and more recently—the leukocyte receptor tyrosine kinase (LTK) gene, expanding the number of targetable driver oncogenes in NSCLC (12,13). Interestingly, gene fusions involving these genes are not specific to NSCLC and are also found in other hematological and solid tumors (14). Functional studies have revealed that NSCLC harboring these gene fusions are totally addicted to the oncogenic fusions, and clinical trials have demonstrated the efficacy of targeting these fusions with specific inhibitors (15) (Figure 1).
In lung cancers, the cigarette smoke is recognized as a major exogenous contributor to cancer development because it contains many mutagenic compounds that can interact directly with the DNA and contribute to genomic instability. However, NSCLCs characterized by gene fusions are rarely associated with an history of smoking and the causes of recurrent oncogenic rearrangements are only partially elucidated (4,16). Endogenous and exogenous factors can trigger DNA rearrangements resulting in fusion genes. Indeed, cellular stress, aberrant DNA repair and recombination, spatial proximity are some factors that can trigger DNA rearrangements, as well as exposure to radiation or chemical compounds. In general, oncogenic rearrangements are the result of double-strand DNA breaks (DSBs) at two different loci, which are then aberrantly repaired by non-homologous end joining (NHEJ) due to defects in the DNA damage repair machinery (17,18). Repair of DSBs occurs by homology-dependent and homology-independent recombination mechanisms (17). Homologous recombination (HR) requires the presence of a high-fidelity matching sequence to allow accurate DNA repair without loss of bases and it is usually activated by mammalian cells in late S and G2 phase, due to the availability of a homologous sister chromatid as a template. Conversely, NHEJ does not require the presence of a DNA strand as a template, therefore functioning in all stages of the cell cycle. It consists in the ligation of the two cut DNA-free ends from either a single gene or entirely separate chromosomes and it can result in DNA translocation from one region or chromosome to another (19). Hence, NHEJ is considered as a highly mutagenic (error-prone) method of DSB repair due to alterations of the original sequence at the sites of break joints and the cause of genomic rearrangements. Recent evidence supports alternative mechanisms of DSB repair at critical regions of the genome, such as coding sequences, to protect from DNA damage associated to oxidative stress (20). In 2016, Wei et al. described the transcription-coupled homologous recombination (TC-HR) as an alternative pathway used by non-dividing cells to obtain high fidelity and efficiency repair of DSBs using RNA transcripts as templates in actively transcribed genes. Emerging evidence indicates that RNA-templated TC-HR may be an important mechanism of DSB repair alternative to NHEJ that protects transcribed genome and prevent genomic rearrangements (20).
DNA repair pathways are then essential to prevent the accumulation of DNA lesions and mutations that may promote tumorigenesis, and defects in DNA repair mechanisms can results in genomic rearrangements. DSBs at two different DNA loci has been identified as the most deleterious type of DNA damage, and diverse mechanisms of chromosomal rearrangements have been reported (4,16). Sometimes there are interchromosomal rearrangements where two non-homologous chromosomes exchange some genetic material. This can be reciprocal (reciprocal translocation), with both chromosomes exchanging genetic material, or non-reciprocal (insertion), with just one of the chromosomes receiving genetic material from the other. Intrachromosomal rearrangements are also possible, in which case the genetic material is exchanged, gained or lost within a single chromosome through the processes of inversion, duplication or deletion. Chromothripsis and chromoplexy are other known mechanisms leading to genomic rearrangements, in which fusions arise due to inaccurate reassembling of chromosome fragments after one chromosome, one chromosome region or a few chromosomes shatter in many fragments and produce a large number of fusion genes in a single event (16,21).
In NSCLC, both reciprocal translocations and inversions that result in gene fusions with altered functions have been reported (22). Reciprocal translocations are usually more frequent than inversions in lung cancer. However, oncogenic ALK fusions, which can be the result of both reciprocal translocations and inversions, are by far mostly represented by an inversion on chromosome 2 that generates the oncogenic fusion Echinoderm microtubule associated protein like 4 (EML4)-ALK (23). RET rearrangements, too, are mainly inversions because most RET fusion partners are on the same chromosome (chrom. 10) of the RET gene.
Typically, in LUAD chromosomal rearrangements that generate gene fusions involve genes that encode receptor tyrosine kinases (RTKs) and can be considered as tyrosine kinase fusions (Table 1). In contrast, gene fusions or mutations involving RTKs are rarely found in LUSC (5). Rearrangements of genes that encode serine-threonine kinases or transcription factors have also been identified in LUAD, but more frequently in LUSC (50).
Table 1
| Oncogene | Fusion partners | Chromosomal rearrangement | Frequency (%) | Type of tumor | Refs |
|---|---|---|---|---|---|
| ALK | EML4- | inv(2)(p21;p23) | 3–7 | NSCLC, papillary thyroid carcinoma, breast carcinoma | (23) |
| TFG- | t(2;3)(p23;q21) | CRC, renal cell carcinoma | (24) | ||
| KIF5B- | t(2;10)(p23;p11) | (25) | |||
| KLC1- | t(2;14)(p23;q32) | (26) | |||
| STRN- | t(2;2)(p23;p22) | (27) | |||
| TPR- | t(1;2)(q31.1;p23) | (28) | |||
| HIP1- | t(2;7)(p23;q11.23) | (29) | |||
| MRPS9- | t(2;2)(p23;q12.1) | (30) | |||
| ROS1 | CD74- | t(5;6)(q32;q22) | 1–3 | NSCLC, glioblastoma, ovarian cancer | (31) |
| EZR- | inv(6)(q22q25.3) | cholangiocarcinoma, CRC, gastric adenocarcinoma | (32) | ||
| TPM3- | t(1;6)(q21.2;q22) | (33) | |||
| SDC4- | t(6;20)(q22;q12) | (33) | |||
| SLC34A2- | t(4;6)(q15.2;q22) | (34) | |||
| LRIG3- | t(6;12)(q22;q14.1) | (33) | |||
| RET | KIF5B- | inv(10)(p11.22q11.2) | 1–2 | NSCLC, papillary thyroid carcinoma, CRC | (33) |
| CCD6- | inv(10)(q11.2q21) | (33) | |||
| NCOA4- | inv(10)(q11.21q11.22) | (35) | |||
| TRIM33- | t(1;10)(p13;q11.2) | (36) | |||
| ERC1- | t(10;12)(q11.2;p13) | (37) | |||
| CLIP1- | t(10;12)( q11.2q24.31) | (38) | |||
| NTRK1 | CD74- | t(1;5)(q23;q33.1) | <5 | NSCLC, CRC, papillary thyroid carcinoma, SCC | (39) |
| MPRIP- | t(1;17)(q23; p11) | (39) | |||
| TPM3- | inv(1)(q21.3q23) | (39) | |||
| SQSTM1- | t(1;5)(q23;q35) | (40) | |||
| NTRK2 | TRIM24- | t(7;9)(q33;q21) | (41) | ||
| NTRK3 | ETV6- | t(12;15)(p13.2;q25.3) | (42) | ||
| NRG1 | CD74- | t(5;8)(q33.1;p12) | 0.1–0.4 | Adenocarcinoma, squamous cell lung carcinoma, pancreatic cancer, gallbladder cancer, renal cell carcinoma | (43) |
| SLC3A2- | t(8;11)(p12;q12.3) | (44) | |||
| SDC4- | t(8;20)(p12;q13.12) | (45) | |||
| VAMP2- | t(8;17)(p12;p13.1) | (46) | |||
| ATP1B1 | t(1;8)(q24.2;p12) | (47) | |||
| FGFR1 | BAG4- | t(8;8)(p11.23;p11.23) | <1 | Squamous cell lung carcinoma, cholangiocarcinoma | (48) |
| FGFR2 | SHTN1- | t(10;10)(q25;q26) | (49) | ||
| CIT- | t(10;12)(q26;q24) | (49) | |||
| FGFR3 | TACC3- | del(4)(p16;p16) | (48) | ||
| LTK | CLIP1- | t(12;15)(q24;q15.1) | 0.4 | NSCLC | (13) |
NSCLC, non-small cell lung cancer; ALK, anaplastic lymphoma kinase; ROS1, C-ros oncogene 1; NTRK, neurotrophic tyrosine kinase receptor; CRC, colorectal cancer; SCC, squamous cell carcinoma; NRG1, neuregulin 1; FGFR, fibroblast growth factor receptor; LTK, leukocyte receptor tyrosine kinase.
Common features of oncogenic fusion-driven NSCLC
Tyrosine kinase fusions are characterized by the juxtaposition of the 5' region of an unrelated gene (fusion partner) to the 3' region encoding the kinase domain of the RTK gene (14). At the C-terminus the resulting fusion proteins (chimeric proteins) retain the intracellular kinase domain of the RTKs fused to the dimerization or oligomerization domains of the fusion partner at the N-terminus that leads to ligand-independent signaling. In fact, due to the dimerization/oligomerization domain of the partner, the oncogenic fusion proteins exhibit constitutive tyrosine kinase activity that generates downstream oncogenic signaling and promotes cell proliferation and survival (14) (Figure 2). Interestingly, FGFR fusions are an exception to this rule because the 5' region of the FGFR gene is fused to the 3' of the partner gene and the resulting fusion chimera retains the extracellular, transmembrane and tyrosine kinase domain of FGFR (51). In addition, because the fused TK is under the control of the regulatory regions of the fusion partner, the fusion proteins are sometimes ectopically expressed in a specific cell or tissue, and the level of expression is dependent on the transcriptional activity of the partner gene. For example, ALK fusions are always ectopically expressed in NSCLC because physiological ALK RTK expression is restricted to embryonic development and to some cells of the adult central nervous system (CNS) (23).
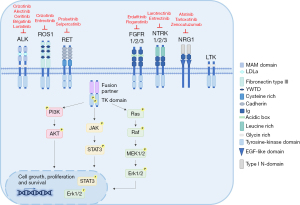
NSCLC harboring oncogenic chimeras share a common biological framework. First, they have a tyrosine kinase domain that is aberrantly activated and drives tumorigenesis. NRG1 fusions are an exception, because NRG1 is not an RTK (see the section on NRG1 in this review). Second, there is a fusion partner that contributes dimerization or oligomerization domains and allows spontaneous dimerization and constitutive activation of the kinase in the absence of the physiological ligand. Indeed, EML4 provides a coiled-coil domain for the dimerization of EML4-ALK fusions as well as the coiled-coil domain containing 6 (CCDC6) protein for the fusion CCD6C-RET (52). In contrast, almost none of the partners in ROS1 fusions feature dimerization or coiled-coil domains, or other dimerization motifs that lead to the constitutive kinase activity of the fusion proteins, therefore a different mechanism of activation can be operating (11). In general, dimerization of the fusion protein is needed for the transformation to take place. Third, different fusion partners have been described for all of the oncogenic fusions derived from different chromosomal rearrangements, including reciprocal translocations and inversions (Table 1). In some cases, only slightly different biological properties have been reported so far, and oncogenic fusions with different partners yield comparable efficacy to targeted therapy. Conversely, some oncogenic fusions display several variants due to different breakpoints in the partner gene resulting in fusion proteins with different regions fused to the kinase domain of the RTK gene. Fusion variants exhibit specific biological and molecular properties and in some cases also different sensitivities to specific TKIs, as reported for EML4-ALK variants (53,54) (see the section on ALK in this review). Fourth, different RTKs can share the same fusion partner, such as kinesin family member 5B (KIF5B) for ALK and RET, or CD74 for ROS1 and NTRK, or CCDC6 for RET and ROS1 but the biological functions are more related to the aberrant activity of the fused kinase rather than the intrinsic biological properties of the fusion partner.
Another important aspect is the subcellular localization of the oncogenic fusion protein which is commonly dependent on the breakpoint in the RTK gene and the physiological localization of the fusion partner. Subcellular localization is important because it can influence downstream signaling. Typically, fusion proteins lose the extracellular region and the transmembrane domain of the RTK. For example, all ALK fusions in NSCLC are cytoplasmic because all fusion partners localize only in the cytoplasm, whereas in ALK-rearranged anaplastic large cell lymphoma (ALCL) NPM-ALK, the most frequent fusion, shows cytoplasmic and nuclear localization due to NPM physiological subcellular localization (55). ROS1 and RET are also commonly cytoplasmic, but in some rearrangements they can retain the transmembrane domain and localize differently (11,52). For example, some ROS1 fusions, SDC4-ROS1 and SLC34A2-ROS1, show endosome-localization with subsequent strong activation of the MAPK pathway, whereas CD74-ROS1 has an endoplasmic reticulum localization (11).
Remarkably, recurrent fusions found in NSCLC have been described in other tumors: ALK, ROS1, RET, NTRK1 and FGFR1/2/3 fusions have been detected in glioblastoma, melanoma, thyroid cancer, breast cancer, bladder cancer, prostate cancer and colorectal cancer (16). In some cases, the same fusion protein has been identified, in other cases different fusions have been reported (14). However, although in different tissues or organs, tumors driven by the same kinase show common or shared molecular mechanisms of tumorigenesis suggesting that they depend more on the activity of the driver oncogene rather than the cell or tissue of origin (8). Indeed, all the tumors harboring recurrent oncogenic fusions are fully addicted to the tyrosine kinase fusions and show high sensitivity to targeted agents (5,7). For example, ALK fusions are the driver oncogenes of both ALCL, specifically NPM-ALK, and NSCLC, specifically EML4-ALK, and both tumors are fully addicted to ALK and fully respond to treatment with ALK TKIs (56). Interestingly, some fusion proteins are found in a specific subset of tumors, although the biological reasons are not fully clarified. For example, NPM-ALK fusion has been identified only in hematological tumors, such as ALCL and diffuse large B cell lymphoma (DLBCL), whereas EML4-ALK has been described only in epithelial tumors (55).
Several studies using in vitro experimental models and in vivo models have clarified the transforming properties of all fusion proteins and clarified which pathways are frequently activated (57). Usually, multiple pathways including RAS/ERK/MAPK, PI3K/AKT and JAK/STAT are commonly activated by RTK fusions to promote and regulate tumor growth and survival (8,57). In addition, functional in vitro and in vivo studies have demonstrated that inhibition of the tyrosine kinase activity of the fusion and the subsequent abolition of downstream signaling leads to cell death and tumor growth arrest (23,58). Accordingly, clinical trials using targeted agents have validated oncogenic fusion proteins as actionable targets for NSCLC (5,15).
Typically, LUADs driven by oncogenic driver mutations are characterized by a low tumor mutational burden (TMB). Notably, high TMB is commonly associated with smoking in lung cancer, instead LUAD harboring actionable driving genomic alterations, such as oncogenic fusions, usually occur in patients with light or never-smoking history. Moreover, the presence of a strong oncogenic driver likely reduces the probability of acquiring additional mutations (5). This has important clinical consequences as high TMB is predictive of the response to immunotherapy with immune checkpoint inhibitors (ICIs) targeting mainly programmed cell death-1 (PD-1) or its ligand PDL-L1. In fact, several clinical trials have demonstrated that NSCLC patients with advanced stage disease in the absence of a driver oncogene and high TMB greatly benefit from ICIs treatment either alone or combined with chemotherapy (59). In contrast the use of ICIs to treat oncogene-addicted NSCLC is still controversial, as it is not clear whether ICIs can be beneficial for these patients alone or in addition to TKI treatment (60). In ALK-driven NSCLC with metastatic disease, ICIs have shown low activity as monotherapies. Indeed, different clinical trials and studies (phase II ATLANTIC trial, phase III OAK trial or IMMUNOTARGET study) have reported little or no response in patients treated with ICI alone although conclusions were limited by the small number of patients enrolled in these studies. Combination with ALK-TKIs resulted in greater toxicity and no additional benefits were observed regardless of PD-L1 expression (59). Similar results have been reported for RET-rearranged NSCLC mainly based on retrospective studies (59). Interestingly, for ROS1 a retrospective analysis reported responses to ICIs administered as single agent, but there was also a correlation with strong PD-L1 expression (61). For other driver mutations, such as NTRK or NRG1, no data are currently available. Overall, further studies with larger cohort of patients are necessary to clearly define the efficacy of ICIs in patients with oncogene-driven NSCLC.
Oncogenic fusions in NSCLC
ALK
The ALK gene encodes for a receptor tyrosine kinase receptor whose physiological functions are still under investigation (uncertain) (55). In 2007, two different studies reported ALK rearrangements in NSCLC (23,24). The frequency of ALK rearrangements in NSCLC is approximately 5–6% and most frequently occurs in LUADs with mucinous cribriform pattern. Different ALK fusions have been reported thus far in NSCLC and include EML4, TRK-fused gene (TFG), KIF5B, kinesin light chain 1 (KLC1), striatin (STRN), huntingtin interacting protein 1 (HIP1), translocated promoter region (TPR) and more recently MRPS9 (8,23,25,26,29,33) (Table 1). The EML4 gene is the predominant fusion partner and several variants of EML4-ALK have been described in NSCLC patients due to different breakpoints in the EML4 gene (occurring at exons 2, 6, 13, 14, 15, 18, 20) (53,62). As described above, all ALK fusions retain the entire intracytoplasmic portion of ALK containing the kinase domain. Indeed, ALK fusions occur at exon 20 of ALK with few exceptions where some studies reported EML4-ALK fusions occurring at exons 18 or 19 of ALK (63-65). Interestingly, all ALK fusions in NSCLC are localized in the cytoplasm and have transforming properties as demonstrated by several in vitro and in vivo studies (62). They promote cell proliferation and survival mainly through the Ras/MAPK/ERK and PI3K/AKT downstream pathways in NSCLC (66). In addition, ALK-driven NSCLC is addicted to ALK tyrosine activity, and experimental and clinical evidence has validated ALK fusions as actionable therapeutic targets (7). Indeed, crizotinib, a first generation ALK TKI was the first ALK TKI approved in 2011 for the treatment of ALK-driven NSCLC (9) as different clinical trials proved that patients treated with crizotinib had improved ORRs and median PFS compared to those treated with standard chemotherapy (67,68). Other second (ceritinib, brigatinib, alectinib) and third (lorlatinib) generation ALK TKIs have been developed and have shown clinical efficacy in treating ALK-rearranged NSCLC (7). Specifically, alectinib not only increased the ORRs and median PFS of untreated patients compared to crizotinib but also showed the ability to cross the blood-brain barrier and effectively target brain metastasis (69). In 2020, alectinib was approved as a first-line treatment option for advanced ALK-rearranged NSCLC and is currently widely used as a first-line therapy for ALK-rearranged NSCLC (70,71).
Although treatment with ALK TKIs has shown a remarkable clinical response, the onset of acquired resistance to ALK TKIs remains a major challenge for the treatment of ALK-driven NSCLC. Several mechanisms of acquired resistance to ALK TKIs have been reported in NSCLC, including secondary ALK mutations that are associated with resistance to all currently available ALK TKIs (5). Mechanisms of resistance to TKIs are discussed in more detail in a separate paragraph of this review. To date, lorlatinib has proven to be effective against all ALK acquired mutations in vitro and has shown efficacy in patients who have become resistant to currently available TKIs, including second-generation ALK TKIs in ALK-positive NSCLC (72). Nonetheless, ALK resistance mutations have been described for lorlatinib (73). However, in a recent study gilteritinib, a new agent approved for the treatment of relapsed/refractory FLT3- positive acute leukemia, displayed potent activity against ALK-TKI resistant mutations and to note also efficacy against ROS1 and NTRK fusions (73).
Interestingly, experimental observations reported that EML4-ALK variants as well as different fusion partners can influence drug sensitivity to ALK TKIs (53,54). Different recent clinical studies revealed differential responses to crizotinib based on the ALK variant, where variant 3 (E6;A20) was less responsive than variant 1 (E13;A20) to ALK inhibition (54,63). Same results have been described in those patients carrying the EML4-ALK variant with the fusion at exon 18 of ALK (E6;A18) (63). In addition, they also observed a significant association between ALK variants and the development of ALK resistance mutations suggesting a link between variants and clinical outcome (54). However, further studies are necessary to expand these observations and eventually establish the selection of ALK TKIs for ALK-rearranged NSCLC treatment based on ALK variants (74).
A new class of compounds has been recently developed aimed at overcoming or delaying ALK TKI resistance (10). These are small molecule degraders that can induce degradation of the target through the proteasome. They are based on proteolysis-targeting chimera (PROTAC) technology by linking ALK TKI analogues to the cereblon ligand pomalidomide. To date, ceritinib, brigatinib and alectinib-based PROTACs have been developed and have shown anti-ALK activity by inducing ALK protein degradation and therefore potent anti-proliferative activity in the ALK-dependent cell lines (75-77). Although preliminary, these data indicate that degraders can represent a promising new option for targeted ALK therapies.
ROS1
ROS1 is a receptor tyrosine kinase that shares a high homology with the insulin receptor superfamily, and it is normally expressed in several organs, although its physiological function is still unclear.
In 2007, ROS1 gene rearrangements were identified in NSCLC tumors for the first time (24). Rikova and colleagues performed a phosphoproteomic screening of both NSCLC cell lines and tumors to identify which kinases showed higher signaling compared to their average activity in non-cancerous samples: RT-PCR of these transcripts revealed two fusion proteins, SLC34A2-ROS1 and CD74-ROS1, already described in glioblastomas but never in NSCLCs.
ROS1 fusions are mainly generated by interchromosomal events and three major breakpoints have been identified proximal to the kinase domain at exons 32, 34 or 35 (33). All rearrangements generate a fusion protein that retains an intact ROS1 intracellular tyrosine kinase domain, while only the exon 32 breakpoint allows to also the maintenance of the transmembrane domain. In contrast to other oncogenic fusions, not all ROS1 fusion partners harbor coiled-coil domains indicating that homodimerization might not be necessary for the oncogenic kinase activation. It is possible that conformational changes induced by the fusion and localization to novel subcellular locations might be sufficient to produce a constitutively active kinase that promotes signal transduction via PI3K/AKT, STAT and MAPK pathways (11,78). Several additional ROS1 fusion partners have been identified throughout the years and they have all been proven to have transforming capabilities both in vitro and in vivo. The most common genes include SLC34A2, CD74, TPM3, SDC4 and EZR, which account for 0.1–1.7% of NSCLC tumors (33).
Given the structural similarity with ALK, ROS1 fusions were initially treated with the ALK-TKI crizotinib, which efficiently blocked cancer progression by inhibiting ROS1 activity and an ORR of the 72% was reported for this cohort of patients (79). Crizotinib was FDA approved for the treatment of ROS1 rearranged NSCLC in 2016. In addition to crizotinib, a more potent ALK TKI, lorlatinib, was assessed in a phase I/II clinical trial in ROS1 rearranged NSCLC patients (80). Lorlatinib showed similar efficacy to crizotinib or other ROS1 TKIs in treatment-naïve patients but was also active against some forms of crizotinib-resistance. Recently, the IFCT-1803 LORLATU study confirmed lorlatinib as a major treatment for advanced refractory ROS1 NSCLC (81). In 2019, entrectinib, a multi-kinase inhibitor active against ROS1 and TRK A, B and C, was approved by the FDA for adults with metastatic NSCLC harboring ROS1 rearrangements. The drug was tested in three clinical studies and showed more potent activity and higher survival rates than crizotinib and the ability to effectively cross the blood-brain barrier and target CNS metastasis (82).
In addition, new ROS1 specific inhibitors, such as repotrectinib and taletrectinib are currently undergoing early phase studies for ROS1-positive NSCLC.
Rearranged during transfection (RET)
The RET gene encodes for a receptor tyrosine kinase with well-established proto-oncogene properties. First identified in CCDC6-RET and NCOA4-RET rearrangements in papillary thyroid carcinoma, in 2012 several groups have simultaneously reported the discovery of a novel fusion transcript in NSCLC between RET and the KIF5B gene that was able to drive oncogenic transformation through the constitutive activation of the RET tyrosine kinase (33). Subsequently, several other fusion partners have been identified using different screening strategies, including CCDC6, NCOA4, TRIM33, ERC1 and CLIP1, mainly in the adenocarcinoma subtype of NSCLC (83). However, the most common RET fusions are KIF5B-RET in approximately 70–90%, followed by CCDC6-RET in 10–25% of RET-rearranged NSCLC (52,84).
The RET gene is located on chromosome 10 where most of its identified fusion partners are located, thus pericentric and paracentric inversions are the main mechanisms of RET rearrangements. Sequencing of breakpoint genomic segments revealed a 2 kB breakpoint susceptible region spanning exon 11 to intron 11. In fact, different variants have been reported for each RET-fusion protein depending on the breakpoint in the RET gene. Generally, all resulting fusion proteins harbor the 3' RET intracellular tyrosine kinase domain while only a few that break at exon 11 retain the transmembrane domain. Moreover, similar to ALK rearrangements, RET fusion partners include a coiled-coil dimerization domain to promote ligand-independent constitutive activation of the kinase (85). Upon activation, the kinase is phosphorylated on multiple tyrosine residues, allowing the direct or adaptor-mediated activation of a wide range of downstream signaling pathways that promote cell growth, proliferation and survival (86).
Due to the high structural homology between RET and other tyrosine kinases, different multi-kinase inhibitors have been evaluated in RET-rearranged NSCLC, such as vandetanib, cabozantinib and lenvatinib, with contradicting results mainly due to treatment toxicities (83). In addition, RET-specific inhibitors have been developed and are currently standard therapeutic regimens for the treatment of RET-rearranged NSCLC patients. Selpercatinib (LOXO-292) is a highly specific small molecule RET-inhibitor and tested in the phase I–II open-label clinical trial LIBRETTO-001. Selpercatinib was then approved in 2020 by the FDA as a first-line therapy for RET-rearranged NSCLC (38). Pralsetinib (BLU-667) is a potent RET inhibitor that showed a stronger in vitro activity against common RET fusions than multi-kinase inhibitors. Pralsetinib has been recently FDA approved for RET-rearranged advanced NSCLC patients given the prosing data from the ARROW trial (38).
NTRK
Oncogenic fusions involving TRK family members, namely TrkA, TrkB and TrkC, which are encoded by the NTRK1, NTRK2 and NTRK3 genes, respectively, have been described in various malignancies, including colorectal cancer, papillary thyroid carcinoma and a subset of NSCLCs. Despite the variety of fusion partners, all resulting chimeras retain the tyrosine kinase domain of NTRK which is constitutively activated by the fusion-partner oligomerization domains and leads to alterations in MAPK, PI3K/AKT and PCL-γ downstream pathways.
The first evidence of NTRK rearrangements in lung cancer was reported in 2013 when Vaishnavi and colleagues identified two novel in-frame fusion events between NTRK1 and the CD74 and MPRIP genes while screening a panel of 36 LUAD samples with no other known genetic alterations (39). In this study, the TPM53-NTRK1 fusion characteristic of colorectal cancer was also described in one of the NSCLC patients.
In addition to NTRK1, other members of the NTRK family are involved in lung cancer: TRIM24-NTRK2 fusion was identified in 2014 by Stransky and colleagues when searching novel gene fusions in the dataset from TCGA (41), while ETV6-NTRK3 fusion has been recently characterized by von der Thüsen et al. (42) in a case report of a patient with a novel primary peribronchial mucinous adenocarcinoma which they named “ETV6-NTRK3 translocation-associated low-grade mucinous bronchial adenocarcinoma” (42).
Preclinical data have demonstrated that TRKs can be efficiently targeted by available TKIs, and several drugs are currently undergoing different phases of clinical trials. Of note, larotrectinib and entrectinib have received approval and breakthrough status from FDA for the treatment of pediatric and adult solid tumors harboring NTRK fusions. Interestingly, in these tumors acquired resistance to TRK inhibitors have been already reported (87).
NRG1
NRG1 fusions have been identified as potentially actionable oncogenic targets in approximately 0,2% of NSCLCs, prevalently in invasive mucinous adenocarcinomas (IMAs) (88).
The gene, located on chromosome 8p12, encodes for a protein-ligand for the ERBB family receptors, specifically ERBB3 and ERBB4. Translocated NRG1 show several fusion partners that vary widely across different tumor types, influencing the biology of the resulting fusion protein (47). The most common fusion partner within NSCLCs is CD74 identified in 2014 during a transcriptome sequencing screening of an Asian cohort of LUAD patients negative for known oncogenic alterations (43). Other fusion partners include SLC3A2, RBPMS, ATP1B1, SDC4 and VAMP2 and all fusions are mutually exclusive with other chromosomal rearrangements (88).
NGR1 genetic rearrangements are structurally similar to those involving RTKs such as ALK, ROS1 and RET, but they do not directly trigger a constitutively active tyrosine kinase. The CD74-NRG1 fusion causes extracellular overexpression of the EGF-like domain of NRG1, which is able to bind ErbB3 that in turn dimerizes with ErbB2, leading to aberrant activation of downstream signaling pathways, including MAPK and PI3K-AKT and stimulation of oncogenic growth.
To date, there are no NRG1 target-specific drugs and no approved therapies for NRG1 rearranged cancers. However, promising responses have been reported after in vitro and in vivo treatment with ErbB targeting agents such as afatinib and tarloxotinib, both irreversible pan-ErbB inhibitors, and zenocutuzumab (MCLA-128), an ErbB2/ErbB3 bispecific antibody. Prospective clinical studies and basket trials are also currently ongoing to better define the role and the efficacy of targeted therapy for NRG1-rearranged NSCLC patients (89).
FGFR
FGFR aberrations are implicated in the development and progression of a wide array of cancer types, including NSCLC. In lung cancer the incidence of FGFR fusions is higher in LUSC than in LUAD (2% vs. 3%) (90). The FGFR family is composed of four members, FGFR1-4, that are active tyrosine kinases and a fifth protein, FGFR5, that lacks the TK domain. In lung cancer the first FGFR rearrangement was identified in 2012 by Seo and colleagues involving the FGFR2 and Citron Rho-interacting kinase (CIT) genes (49). Through genomic profiling several fusion genes between FGFR1, FGFR2 or FGFR3 and different partner genes have been discovered in NSCLC samples, where FGFR3-TACC3 fusion is the most represented.
Interestingly, FGFR-TACC3 fusions have also been found in tumor tissue from patients with EGFR-driven NSCLC previously treated with EGFR TKIs. Additional analyses confirmed that this alteration was acquired during treatment (51). Therefore, these data support FGFR rearrangements not only as a driver oncogene in NSCLC but also as a mechanism of acquired resistance to targeted therapy (51).
Targeted therapy options involving selective and non-selective FGFR inhibitors are under investigation for FGFR-rearranged lung cancer patients. Non-selective inhibitors are multitarget TKIs, i.e., ponatinib, which is FDA approved for the treatment of chronic myeloid leukemia and acute lymphoblastic leukemia, is currently under evaluation in multiple clinical trials for NSCLC patients harboring FGFR1 rearrangements, but the results have not yet reported. In contrast, selective inhibitors are drugs developed to specifically target FGFR RTKs and ongoing clinical trials are investigating the efficacy of erdafitinib and rogaratinib, both oral pan-FGFR inhibitors, in FGFR-rearranged NSCLC patients (91).
LTK
A novel in-frame fusion transcript between CLIP, on chromosome 12q24, and LTK, on chromosome 15q15 that generates the CLIP1-LTK fusion has been newly discovered in a NSCLC patient through whole transcriptome sequencing of 75 NSCLC samples from the LC-SCRUM-Asia cohort (13). Further screening by RT-PCR revealed the CLIP1-LTK transcript in two additional patients of this Asian cohort.
LTK is an RTK that belongs to the ALK/LTK subfamily within the insulin receptor superfamily. Although its biological role remains unclear, it is known that LTK is able to signal through RAS/MAPK and PI3K/AKT (92). In addition, LTK is frequently overexpressed in several malignancies, including NSCLC, where it is associated with a higher risk of metastasis (93). However, for the first time an LTK fusion was described and proven to drive transformation both in vitro and in vivo (13). As described for other oncogenic fusion proteins, the coiled-coil domain of CLIP1 leads to dimerization and constitutive activation of the kinase domain of LTK.
Since LTK shares an 80% kinase domain identity with ALK, pre-clinical evidence showed that all ALK TKIs had variable degree of efficacy against CLIP1-LTK fusion. In addition, in one NSCLC patient harboring the fusion and refractory to chemotherapy treated with a standard dosage of lorlatinib had a dramatic clinical response. LTK fusions can therefore represent a new therapeutically actionable oncogenic driver in NSCLC although further studies are necessary to characterize LTK-fusions in NSCLC.
How to diagnose gene fusions in NSCLC
The individualized approach for the treatment of NSCLC patients has made early identification of tumor genotype essential to prescribe the most efficacious therapy available. As well as other genetic alterations, tyrosine kinase fusions represent strong predictive biomarkers in NSCLC and can be identified by several diagnostic techniques.
Indeed, in routine clinical practice it is now mandatory that patients with newly diagnosed advanced NSCLC are tested for specific genetic alterations, including rearrangements of ALK, ROS1, RET and NTRK genes (94,95). Since several genetic tests are required and in many cases the amount of available tumor sample is insufficient to perform multiple molecular assays, such as FISH or immunohistochemistry (IHC), simultaneous screening through NGS is replacing the sequential single fusion testing for the molecular diagnosis of NSCLC. Several assays have been developed to diagnose gene translocations in NSCLC and are nicely reviewed elsewhere (95). However, the choice of using single gene testing or NGS will change accordingly with costs, laboratory environment, specific patient needs, the tissue biopsy quality and availability, and the source of tumor specimen. To note, the reliability of molecular tests is largely dependent on the source of tumor specimen and on the many tissue-handling steps of the preanalytical phase. Interestingly, for fusion gene assessment RNA-based NGS analysis is concurrently emerging as a parallel strategy for screening or confirming the presence of fusion genes, and more likely will be implemented in the future (1,96). Indeed, DNA-based assays sometimes lack information about which exons are involved in the rearrangement and do not provide information about the expressed fusion transcript. In addition, RNA-based NGS assays are more sensitive than DNA-based approaches and can provide useful information for targeted therapy, but they show some limitations. Fusion genes can occur with different fusion partners and some RNA-based NGS platforms are not designed to detect all the different fusions (95).
Recent studies have reported that lung cancer cells shed tumor DNA into the bloodstream opening to the use of liquid biopsy to genotype advanced NSCLCs, with advantages in cost-convenience, low invasiveness of the procedure and faster results. There are two sources of tumor DNA in the circulation, cell-free circulating DNA (ctDNA) and circulating tumor cells (CTCs). Although the mechanisms are still unclear, it has been demonstrated that the amount of detectable ctDNA depends to the overall tumor burden and it usually is sufficient to detect known oncogenic driver mutations in the majority of lung cancer patients with one or more metastatic sites (97).
The Blood First Assay Screening trial (BFAST- NCT03178552) is an ongoing global, multicohort study designed to evaluate the efficiency and safety of targeted therapies in patients with treatment-naïve advanced or metastatic NSCLC who were screened for specific genetic alterations using only blood-based NGS. Currently, six cohorts have been enrolled, including ALK-positive, RET-positive and ROS1-positive NSCLC patients (98). In a first data report from the ALK cohort, retrospective testing of ALEX (BO28984) plasma samples was used to investigate blood-tissue marker concordance, comparing IHC ALK-positive diagnosis with blood-based testing. Results found 70.5% concordance between IHC and blood-based NGS, suggesting that both diagnostic strategies can be used to select patients for targeted therapy. In addition, BFAST primary data demonstrated that ctDNA analysis represents a less invasive diagnostic tool useful for patients with insufficient tumor sample for FISH and IHC ALK rearrangement testing (98).
Following these trials, between 2018 and 2019, the U.S. FDA granted breakthrough designation to three commercial NGS-based liquid biopsies that include all the genes recommended as first-line testing in the U.S. and in most European Countries (94).
Mechanisms of resistance to TKIs in oncogenic fusion-driven NSCLC
TKIs have changed the clinical care of NSCLC, replacing the use of chemotherapy as initial treatment for many patients. However, despite the initial benefits of targeted therapy, long-term disease control in NSCLC patients is frequently limited by acquired drug resistance that drives disease progression and tumor relapse. Resistance can be also the consequence of tumor heterogeneity that makes tumors unresponsive to therapy from the beginning (99,100).
Recent evidence points out that oncogenic driver fusion-defined subgroups of NSCLCs are also clinically heterogeneous; indeed, beside the main oncogenic fusions, cancer cells can simultaneously harbor different genetic aberrations in several oncogenic pathways, leading to variable clinical behavior and altered sensitivity to anticancer therapies. In general, overlap with alterations in other targetable oncogenes (i.e., RAS, EGFR, ALK) in NSCLC is rarely reported (101). However, among advanced-stage TKI-rearranged NSCLCs, coexisting mutations in TP53 have not been frequently observed, whereas alterations of CDKN2A and CDKN2B co-occur at high rates in ALK-, ROS1- and RET-rearranged NSCLC and are correlated with adverse prognosis (21,101). To note, the presence of TP53 mutations might adversely impact the survival of NSCLC patients with ALK and ROS1 fusions, leading to genome instability and thus accelerating the onset of different mechanisms of resistance to targeted therapy (102,103).
Acquired resistance is mediated by diverse mechanisms, so called “on target”, including mutations in the drug target or amplification of the target and “off target” mechanisms, including activation of bypass signaling pathways, mutations in downstream effectors, and, in some cases, state transformation [i.e., from NSCLC to small cell lung cancer (SCLC)], that ultimately result in maintenance of proliferative signaling despite the presence of the TKI.
In ALK-rearranged NSCLC, resistance to TKIs is mainly caused by ALK secondary mutations: the most common are the gatekeeper mutations L1196M and G1269A, that interfere with the drug binding, and the G1202R mutation, that reduces the affinity to the ALK TKI. Interestingly, the G1202 mutation is also refractory to second generation ALK-TKIs but can be overcome by the third generation ALK-TKI lorlatinib. On-target secondary mutations have been reported for other kinase-rearranged lung cancers: in ROS1-crizotinib resistant patients, the G2032R mutation within the ATP-binding pocket is the most frequently diffused together with D2033N, L2026M, and S1986F/Y mutations, while NTRK1 G595R and NTRK3 G623R mutations are causative of resistance to larotrectinib in NTRK-positive NSCLCs (72,104-107).
Activation of alternative survival signaling pathways has also been indicated as a major mechanism of resistance. In this case, cancer cells switch their dependence to other RTKs bypassing the inhibition to the drug target. In approximately 30% of crizotinib relapsing EML4-ALK patients resistance is mediated by either EGFR activation or c-KIT amplification whereas MET amplification has been reported in alectinib relapsing ALK+NSCLC (104). Other examples of alternative signaling pathways activation are BRAF and KRAS activation and MET-amplification in ROS1-rearranged TKI-resistant NSCLC and acquired MET or KRAS amplifications in 15% of RET-fusion positive resistant lung cancers (87).
In addition, histologic transformation from adenocarcinoma to SCLC, described for the first time in a case of EGFR-mutant LUAD reported by Zakowski et al. in 2006 (108), has now emerged as an important mechanism of resistance in TKI-resistant adenocarcinomas with ALK and ROS1 oncogenic fusions (104,109). However, the molecular mechanisms remain to be elucidated as well as the possible therapeutic strategies that could be adopted to overcome this type of resistance.
Conclusions
NSCLC was previously thought quite as a relatively homogeneous tumor entity, but with the advent of high throughput genetic screening and the identification of recurrent oncogenic lesions is now recognized in distinct clinical entities. In addition, the impressive therapeutic clinical response obtained in patients that harbor oncogenic fusion proteins using specific inhibitors (small molecule drugs) has validated oncogenic chimeras as therapeutic targets. Indeed, the molecular identification of targetable driver fusions such as ALK, ROS1 or RET are currently included in the diagnostic process and considered fundamental to stratify NSCLC patients and to determine the optimal targeted therapy. In addition, next generation sequencing technologies are currently standard practice to define the molecular status of NSCLC at diagnosis that provide key information to predict response to therapy.
However, there are still ongoing challenges. Approximately 25–30% of NSCLC are orphan for driver oncogenes. The identification of actionable genetic alterations could help a better stratification of NSCLC patients and allow individualized therapy. Elucidation of the molecular mechanisms that lead to oncogenic rearrangements could help defining risk factors or developing preventive methods to reduce the incidence of these malignancies.
Acknowledgments
Funding: This work was supported by Associazione Italiana Ricerca sul Cancro (AIRC) (IG 2019 - ID.23146 to CV).
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Fabrizio Tabbò, Umberto Malapelle, Maria Lucia Reale and Angela Listì) for the series “How to Detect and Treat NSCLC Patients with Oncogenic Fusions” published in Precision Cancer Medicine. The article has undergone external peer review.
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at https://pcm.amegroups.com/article/view/10.21037/pcm-22-7/coif). The series “How to Detect and Treat NSCLC Patients with Oncogenic Fusions” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Barr FG. Fusion genes in solid tumors: the possibilities and the pitfalls. Expert Rev Mol Diagn 2016;16:921-3. [Crossref] [PubMed]
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511:543-50. [Crossref] [PubMed]
- Mertens F, Johansson B, Fioretos T, et al. The emerging complexity of gene fusions in cancer. Nat Rev Cancer 2015;15:371-81. [Crossref] [PubMed]
- Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature 2018;553:446-54. [Crossref] [PubMed]
- Chevallier M, Borgeaud M, Addeo A, et al. Oncogenic driver mutations in non-small cell lung cancer: Past, present and future. World J Clin Oncol 2021;12:217-37. [Crossref] [PubMed]
- Thai AA, Solomon BJ, Sequist LV, et al. Lung cancer. Lancet 2021;398:535-54. [Crossref] [PubMed]
- Tabbò F, Pizzi M, Kyriakides PW, et al. Oncogenic kinase fusions: an evolving arena with innovative clinical opportunities. Oncotarget 2016;7:25064-86. [Crossref] [PubMed]
- Kazandjian D, Blumenthal GM, Chen HY, et al. FDA approval summary: crizotinib for the treatment of metastatic non-small cell lung cancer with anaplastic lymphoma kinase rearrangements. Oncologist 2014;19:e5-11. [Crossref] [PubMed]
- Cohen P, Cross D, Jänne PA. Kinase drug discovery 20 years after imatinib: progress and future directions. Nat Rev Drug Discov 2021;20:551-69. [Crossref] [PubMed]
- Drilon A, Jenkins C, Iyer S, et al. ROS1-dependent cancers - biology, diagnostics and therapeutics. Nat Rev Clin Oncol 2021;18:35-55. [Crossref] [PubMed]
- Kohno T, Nakaoku T, Tsuta K, et al. Beyond ALK-RET, ROS1 and other oncogene fusions in lung cancer. Transl Lung Cancer Res 2015;4:156-64. [PubMed]
- Izumi H, Matsumoto S, Liu J, et al. The CLIP1-LTK fusion is an oncogenic driver in non-small-cell lung cancer. Nature 2021;600:319-23. [Crossref] [PubMed]
- Shaw AT, Hsu PP, Awad MM, et al. Tyrosine kinase gene rearrangements in epithelial malignancies. Nat Rev Cancer 2013;13:772-87. [Crossref] [PubMed]
- Reck M, Rabe KF. Precision Diagnosis and Treatment for Advanced Non-Small-Cell Lung Cancer. N Engl J Med 2017;377:849-61. [Crossref] [PubMed]
- Tuna M, Amos CI, Mills GB. Molecular mechanisms and pathobiology of oncogenic fusion transcripts in epithelial tumors. Oncotarget 2019;10:2095-111. [Crossref] [PubMed]
- Richardson C, Jasin M. Frequent chromosomal translocations induced by DNA double-strand breaks. Nature 2000;405:697-700. [Crossref] [PubMed]
- Mani RS, Chinnaiyan AM. Triggers for genomic rearrangements: insights into genomic, cellular and environmental influences. Nat Rev Genet 2010;11:819-29. [Crossref] [PubMed]
- Burgess JT, Rose M, Boucher D, et al. The Therapeutic Potential of DNA Damage Repair Pathways and Genomic Stability in Lung Cancer. Front Oncol 2020;10:1256. [Crossref] [PubMed]
- Wei L, Levine AS, Lan L. Transcription-coupled homologous recombination after oxidative damage. DNA Repair (Amst) 2016;44:76-80. [Crossref] [PubMed]
- Lee JJ, Park S, Park H, et al. Tracing Oncogene Rearrangements in the Mutational History of Lung Adenocarcinoma. Cell 2019;177:1842-1857.e21. [Crossref] [PubMed]
- Li Y, Roberts ND, Wala JA, et al. Patterns of somatic structural variation in human cancer genomes. Nature 2020;578:112-21. [Crossref] [PubMed]
- Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 2007;448:561-6. [Crossref] [PubMed]
- Rikova K, Guo A, Zeng Q, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell 2007;131:1190-203. [Crossref] [PubMed]
- Takeuchi K, Choi YL, Togashi Y, et al. KIF5B-ALK, a novel fusion oncokinase identified by an immunohistochemistry-based diagnostic system for ALK-positive lung cancer. Clin Cancer Res 2009;15:3143-9. [Crossref] [PubMed]
- Togashi Y, Soda M, Sakata S, et al. KLC1-ALK: a novel fusion in lung cancer identified using a formalin-fixed paraffin-embedded tissue only. PLoS One 2012;7:e31323. [Crossref] [PubMed]
- Nakanishi Y, Masuda S, Iida Y, et al. Case Report of Non-Small Cell Lung Cancer with STRN-ALK Translocation: A Nonresponder to Alectinib. J Thorac Oncol 2017;12:e202-4. [Crossref] [PubMed]
- Choi YL, Lira ME, Hong M, et al. A novel fusion of TPR and ALK in lung adenocarcinoma. J Thorac Oncol 2014;9:563-6. [Crossref] [PubMed]
- Ou SH, Klempner SJ, Greenbowe JR, et al. Identification of a novel HIP1-ALK fusion variant in Non-Small-Cell Lung Cancer (NSCLC) and discovery of ALK I1171 (I1171N/S) mutations in two ALK-rearranged NSCLC patients with resistance to Alectinib. J Thorac Oncol 2014;9:1821-5. [Crossref] [PubMed]
- Zhou H, Xu B, Xu J, et al. Novel MRPS9-ALK Fusion Mutation in a Lung Adenocarcinoma Patient: A Case Report. Front Oncol 2021;11:670907. [Crossref] [PubMed]
- Bergethon K, Shaw AT, Ou SH, et al. ROS1 rearrangements define a unique molecular class of lung cancers. J Clin Oncol 2012;30:863-70. [Crossref] [PubMed]
- Arai Y, Totoki Y, Takahashi H, et al. Mouse model for ROS1-rearranged lung cancer. PLoS One 2013;8:e56010. [Crossref] [PubMed]
- Takeuchi K, Soda M, Togashi Y, et al. RET, ROS1 and ALK fusions in lung cancer. Nat Med 2012;18:378-81. [Crossref] [PubMed]
- Davies KD, Le AT, Theodoro MF, et al. Identifying and targeting ROS1 gene fusions in non-small cell lung cancer. Clin Cancer Res 2012;18:4570-9. [Crossref] [PubMed]
- Wang R, Hu H, Pan Y, et al. RET fusions define a unique molecular and clinicopathologic subtype of non-small-cell lung cancer. J Clin Oncol 2012;30:4352-9. [Crossref] [PubMed]
- Drilon A, Wang L, Hasanovic A, et al. Response to Cabozantinib in patients with RET fusion-positive lung adenocarcinomas. Cancer Discov 2013;3:630-5. [Crossref] [PubMed]
- Drilon A, Rekhtman N, Arcila M, et al. Cabozantinib in patients with advanced RET-rearranged non-small-cell lung cancer: an open-label, single-centre, phase 2, single-arm trial. Lancet Oncol 2016;17:1653-60. [Crossref] [PubMed]
- Drusbosky LM, Rodriguez E, Dawar R, et al. Therapeutic strategies in RET gene rearranged non-small cell lung cancer. J Hematol Oncol 2021;14:50. [Crossref] [PubMed]
- Vaishnavi A, Capelletti M, Le AT, et al. Oncogenic and drug-sensitive NTRK1 rearrangements in lung cancer. Nat Med 2013;19:1469-72. [Crossref] [PubMed]
- Farago AF, Le LP, Zheng Z, et al. Durable Clinical Response to Entrectinib in NTRK1-Rearranged Non-Small Cell Lung Cancer. J Thorac Oncol 2015;10:1670-4. [Crossref] [PubMed]
- Stransky N, Cerami E, Schalm S, et al. The landscape of kinase fusions in cancer. Nat Commun 2014;5:4846. [Crossref] [PubMed]
- von der Thüsen JH, Dumoulin DW, Maat APWM, et al. ETV6-NTRK3 translocation-associated low-grade mucinous bronchial adenocarcinoma: A novel bronchial salivary gland-type non-small cell lung cancer subtype. Lung Cancer 2021;156:72-5. [Crossref] [PubMed]
- Fernandez-Cuesta L, Plenker D, Osada H, et al. CD74-NRG1 fusions in lung adenocarcinoma. Cancer Discov 2014;4:415-22. [Crossref] [PubMed]
- Nakaoku T, Tsuta K, Ichikawa H, et al. Druggable oncogene fusions in invasive mucinous lung adenocarcinoma. Clin Cancer Res 2014;20:3087-93. [Crossref] [PubMed]
- Dhanasekaran SM, Balbin OA, Chen G, et al. Transcriptome meta-analysis of lung cancer reveals recurrent aberrations in NRG1 and Hippo pathway genes. Nat Commun 2014;5:5893. [Crossref] [PubMed]
- Jung Y, Yong S, Kim P, et al. VAMP2-NRG1 Fusion Gene is a Novel Oncogenic Driver of Non-Small-Cell Lung Adenocarcinoma. J Thorac Oncol 2015;10:1107-11. [Crossref] [PubMed]
- Jonna S, Feldman RA, Swensen J, et al. Detection of NRG1 Gene Fusions in Solid Tumors. Clin Cancer Res 2019;25:4966-72. [Crossref] [PubMed]
- Wang R, Wang L, Li Y, et al. FGFR1/3 tyrosine kinase fusions define a unique molecular subtype of non-small cell lung cancer. Clin Cancer Res 2014;20:4107-14. [Crossref] [PubMed]
- Seo JS, Ju YS, Lee WC, et al. The transcriptional landscape and mutational profile of lung adenocarcinoma. Genome Res 2012;22:2109-19. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012;489:519-25. [Crossref] [PubMed]
- Qin A, Johnson A, Ross JS, et al. Detection of Known and Novel FGFR Fusions in Non-Small Cell Lung Cancer by Comprehensive Genomic Profiling. J Thorac Oncol 2019;14:54-62. [Crossref] [PubMed]
- Li AY, McCusker MG, Russo A, et al. RET fusions in solid tumors. Cancer Treat Rev 2019;81:101911. [Crossref] [PubMed]
- Childress MA, Himmelberg SM, Chen H, et al. ALK Fusion Partners Impact Response to ALK Inhibition: Differential Effects on Sensitivity, Cellular Phenotypes, and Biochemical Properties. Mol Cancer Res 2018;16:1724-36. [Crossref] [PubMed]
- Lin JJ, Zhu VW, Yoda S, et al. Impact of EML4-ALK Variant on Resistance Mechanisms and Clinical Outcomes in ALK-Positive Lung Cancer. J Clin Oncol 2018;36:1199-206. [Crossref] [PubMed]
- Chiarle R, Voena C, Ambrogio C, et al. The anaplastic lymphoma kinase in the pathogenesis of cancer. Nat Rev Cancer 2008;8:11-23. [Crossref] [PubMed]
- Solomon B, Wilner KD, Shaw AT. Current status of targeted therapy for anaplastic lymphoma kinase-rearranged non-small cell lung cancer. Clin Pharmacol Ther 2014;95:15-23. [Crossref] [PubMed]
- Hallberg B, Palmer RH. Mechanistic insight into ALK receptor tyrosine kinase in human cancer biology. Nat Rev Cancer 2013;13:685-700. [Crossref] [PubMed]
- Soda M, Takada S, Takeuchi K, et al. A mouse model for EML4-ALK-positive lung cancer. Proc Natl Acad Sci U S A 2008;105:19893-7. [Crossref] [PubMed]
- Calles A, Riess JW, Brahmer JR. Checkpoint Blockade in Lung Cancer With Driver Mutation: Choose the Road Wisely. Am Soc Clin Oncol Educ Book 2020;40:372-84. [Crossref] [PubMed]
- Tsakonas G, Ekman S. Oncogene-addicted non-small cell lung cancer and immunotherapy. J Thorac Dis 2018;10:S1547-55. [Crossref] [PubMed]
- Mazieres J, Drilon A, Lusque A, et al. Immune checkpoint inhibitors for patients with advanced lung cancer and oncogenic driver alterations: results from the IMMUNOTARGET registry. Ann Oncol 2019;30:1321-8. [Crossref] [PubMed]
- Sabir SR, Yeoh S, Jackson G, et al. EML4-ALK Variants: Biological and Molecular Properties, and the Implications for Patients. Cancers (Basel) 2017;9:118. [Crossref] [PubMed]
- Anai S, Takeshita M, Ando N, et al. A Case of Lung Adenocarcinoma Resistant to Crizotinib Harboring a Novel EML4-ALK Variant, Exon 6 of EML4 Fused to Exon 18 of ALK. J Thorac Oncol 2016;11:e126-8. [Crossref] [PubMed]
- Liu L, Hou F, Liu Y, et al. A Case of Lung Adenocarcinoma Response to Alectinib Harboring a Rare EML4-ALK Variant, Exon 6 of EML4 Fused to Exon 18 of ALK. J Natl Compr Canc Netw 2021;20:2-6. [Crossref] [PubMed]
- Penzel R, Schirmacher P, Warth A. A novel EML4-ALK variant: exon 6 of EML4 fused to exon 19 of ALK. J Thorac Oncol 2012;7:1198-9. [Crossref] [PubMed]
- Voena C, Di Giacomo F, Panizza E, et al. The EGFR family members sustain the neoplastic phenotype of ALK+ lung adenocarcinoma via EGR1. Oncogenesis 2013;2:e43. [Crossref] [PubMed]
- Shaw AT, Kim DW, Nakagawa K, et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med 2013;368:2385-94. [Crossref] [PubMed]
- Solomon BJ, Mok T, Kim DW, et al. First-line crizotinib versus chemotherapy in ALK-positive lung cancer. N Engl J Med 2014;371:2167-77. [Crossref] [PubMed]
- Gadgeel SM, Gandhi L, Riely GJ, et al. Safety and activity of alectinib against systemic disease and brain metastases in patients with crizotinib-resistant ALK-rearranged non-small-cell lung cancer (AF-002JG): results from the dose-finding portion of a phase 1/2 study. Lancet Oncol 2014;15:1119-28. [Crossref] [PubMed]
- Peters S, Camidge DR, Shaw AT, et al. Alectinib versus Crizotinib in Untreated ALK-Positive Non-Small-Cell Lung Cancer. N Engl J Med 2017;377:829-38. [Crossref] [PubMed]
- Zhou C, Kim SW, Reungwetwattana T, et al. Alectinib versus crizotinib in untreated Asian patients with anaplastic lymphoma kinase-positive non-small-cell lung cancer (ALESIA): a randomised phase 3 study. Lancet Respir Med 2019;7:437-46. [Crossref] [PubMed]
- Shaw AT, Felip E, Bauer TM, et al. Lorlatinib in non-small-cell lung cancer with ALK or ROS1 rearrangement: an international, multicentre, open-label, single-arm first-in-man phase 1 trial. Lancet Oncol 2017;18:1590-9. [Crossref] [PubMed]
- Mizuta H, Okada K, Araki M, et al. Gilteritinib overcomes lorlatinib resistance in ALK-rearranged cancer. Nat Commun 2021;12:1261. [Crossref] [PubMed]
- Christopoulos P, Kirchner M, Endris V, et al. EML4-ALK V3, treatment resistance, and survival: refining the diagnosis of ALK+ NSCLC. J Thorac Dis 2018;10:S1989-91. [Crossref] [PubMed]
- Zhang C, Han XR, Yang X, et al. Proteolysis Targeting Chimeras (PROTACs) of Anaplastic Lymphoma Kinase (ALK). Eur J Med Chem 2018;151:304-14. [Crossref] [PubMed]
- Sun N, Ren C, Kong Y, et al. Development of a Brigatinib degrader (SIAIS117) as a potential treatment for ALK positive cancer resistance. Eur J Med Chem 2020;193:112190. [Crossref] [PubMed]
- Ren C, Sun N, Kong Y, et al. Structure-based discovery of SIAIS001 as an oral bioavailability ALK degrader constructed from Alectinib. Eur J Med Chem 2021;217:113335. [Crossref] [PubMed]
- Gainor JF, Shaw AT. Novel targets in non-small cell lung cancer: ROS1 and RET fusions. Oncologist 2013;18:865-75. [Crossref] [PubMed]
- Shaw AT, Ou SH, Bang YJ, et al. Crizotinib in ROS1-rearranged non-small-cell lung cancer. N Engl J Med 2014;371:1963-71. [Crossref] [PubMed]
- Shaw AT, Solomon BJ, Chiari R, et al. Lorlatinib in advanced ROS1-positive non-small-cell lung cancer: a multicentre, open-label, single-arm, phase 1-2 trial. Lancet Oncol 2019;20:1691-701. [Crossref] [PubMed]
- Girard N, Galland-Girodet S, Avrillon V, et al. Lorlatinib for advanced ROS1+ non-small-cell lung cancer: results of the IFCT-1803 LORLATU study. ESMO Open 2022;7:100418. [Crossref] [PubMed]
- Drilon A, Siena S, Dziadziuszko R, et al. Entrectinib in ROS1 fusion-positive non-small-cell lung cancer: integrated analysis of three phase 1-2 trials. Lancet Oncol 2020;21:261-70. [Crossref] [PubMed]
- Takeuchi K. Discovery Stories of RET Fusions in Lung Cancer: A Mini-Review. Front Physiol 2019;10:216. [Crossref] [PubMed]
- Santoro M, Moccia M, Federico G, et al. RET Gene Fusions in Malignancies of the Thyroid and Other Tissues. Genes (Basel) 2020;11:424. [Crossref] [PubMed]
- Mizukami T, Shiraishi K, Shimada Y, et al. Molecular mechanisms underlying oncogenic RET fusion in lung adenocarcinoma. J Thorac Oncol 2014;9:622-30. [Crossref] [PubMed]
- Mulligan LM. RET revisited: expanding the oncogenic portfolio. Nat Rev Cancer 2014;14:173-86. [Crossref] [PubMed]
- Russo A, Cardona AF, Caglevic C, et al. Overcoming TKI resistance in fusion-driven NSCLC: new generation inhibitors and rationale for combination strategies. Transl Lung Cancer Res 2020;9:2581-98. [Crossref] [PubMed]
- Laskin J, Liu SV, Tolba K, et al. NRG1 fusion-driven tumors: biology, detection, and the therapeutic role of afatinib and other ErbB-targeting agents. Ann Oncol 2020;31:1693-703. [Crossref] [PubMed]
- Liu SV. NRG1 fusions: Biology to therapy. Lung Cancer 2021;158:25-8. [Crossref] [PubMed]
- Helsten T, Elkin S, Arthur E, et al. The FGFR Landscape in Cancer: Analysis of 4,853 Tumors by Next-Generation Sequencing. Clin Cancer Res 2016;22:259-67. [Crossref] [PubMed]
- Pacini L, Jenks AD, Lima NC, et al. Targeting the Fibroblast Growth Factor Receptor (FGFR) Family in Lung Cancer. Cells 2021;10:1154. [Crossref] [PubMed]
- Roll JD, Reuther GW. ALK-activating homologous mutations in LTK induce cellular transformation. PLoS One 2012;7:e31733. [Crossref] [PubMed]
- Müller-Tidow C, Diederichs S, Bulk E, et al. Identification of metastasis-associated receptor tyrosine kinases in non-small cell lung cancer. Cancer Res 2005;65:1778-82. [Crossref] [PubMed]
- Gregg JP, Li T, Yoneda KY. Molecular testing strategies in non-small cell lung cancer: optimizing the diagnostic journey. Transl Lung Cancer Res 2019;8:286-301. [Crossref] [PubMed]
- Kazdal D, Hofman V, Christopoulos P, et al. Fusion-positive non-small cell lung carcinoma: Biological principles, clinical practice, and diagnostic implications. Genes Chromosomes Cancer 2022;61:244-60. [Crossref] [PubMed]
- Lindeman NI, Cagle PT, Aisner DL, et al. Updated Molecular Testing Guideline for the Selection of Lung Cancer Patients for Treatment With Targeted Tyrosine Kinase Inhibitors: Guideline From the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. Arch Pathol Lab Med 2018;142:321-46. [Crossref] [PubMed]
- Sacher AG, Paweletz C, Dahlberg SE, et al. Prospective Validation of Rapid Plasma Genotyping for the Detection of EGFR and KRAS Mutations in Advanced Lung Cancer. JAMA Oncol 2016;2:1014-22. [Crossref] [PubMed]
- Dziadziuszko R, Mok T, Peters S, et al. Blood First Assay Screening Trial (BFAST) in Treatment-Naive Advanced or Metastatic NSCLC: Initial Results of the Phase 2 ALK-Positive Cohort. J Thorac Oncol 2021;16:2040-50. [Crossref] [PubMed]
- Dagogo-Jack I, Shaw AT. Expanding the Roster of ROS1 Inhibitors. J Clin Oncol 2017;35:2595-7. [Crossref] [PubMed]
- Tulpule A, Bivona TG. Acquired Resistance in Lung Cancer. Annu Rev Cancer Biol 2020;4:279-97. [Crossref]
- Skoulidis F, Heymach JV. Co-occurring genomic alterations in non-small-cell lung cancer biology and therapy. Nat Rev Cancer 2019;19:495-509. [Crossref] [PubMed]
- Aisner DL, Sholl LM, Berry LD, et al. The Impact of Smoking and TP53 Mutations in Lung Adenocarcinoma Patients with Targetable Mutations-The Lung Cancer Mutation Consortium (LCMC2). Clin Cancer Res 2018;24:1038-47. [Crossref] [PubMed]
- Xiao D, Deng Q, He D, et al. High Tumor Mutation Burden and DNA Repair Gene Mutations are Associated with Primary Resistance to Crizotinib in ALK-Rearranged Lung Cancer. Onco Targets Ther 2021;14:4809-17. [Crossref] [PubMed]
- Lin JJ, Shaw AT. Resisting Resistance: Targeted Therapies in Lung Cancer. Trends Cancer 2016;2:350-64. [Crossref] [PubMed]
- Gainor JF, Dardaei L, Yoda S, et al. Molecular Mechanisms of Resistance to First- and Second-Generation ALK Inhibitors in ALK-Rearranged Lung Cancer. Cancer Discov 2016;6:1118-33. [Crossref] [PubMed]
- Awad MM, Katayama R, McTigue M, et al. Acquired resistance to crizotinib from a mutation in CD74-ROS1. N Engl J Med 2013;368:2395-401. [Crossref] [PubMed]
- Drilon A, Laetsch TW, Kummar S, et al. Efficacy of Larotrectinib in TRK Fusion-Positive Cancers in Adults and Children. N Engl J Med 2018;378:731-9. [Crossref] [PubMed]
- Zakowski MF, Ladanyi M, Kris MG, et al. EGFR mutations in small-cell lung cancers in patients who have never smoked. N Engl J Med 2006;355:213-5. [Crossref] [PubMed]
- Calabrese F, Pezzuto F, Lunardi F, et al. Morphologic-Molecular Transformation of Oncogene Addicted Non-Small Cell Lung Cancer. Int J Mol Sci 2022;23:4164. [Crossref] [PubMed]
Cite this article as: Di Marco MV, Voena C. Review: biological implications of oncogenic rearrangements in non-small cell lung cancer. Precis Cancer Med 2022;5:26.

