
Current state of the art on the diagnosis and the role of target therapy for treatment of ROS1-rearranged non-small cell lung cancer: a narrative review
Introduction
Background
Non-small cell lung cancer (NSCLC) is currently classified according to histology and to molecular features. Approximately two-thirds of advanced NSCLCs are identified as oncogene-addicted disease, due to the occurrence of driver genetic alterations at the time of diagnosis, including mutations (e.g., EGFR, KRAS, BRAF, MET, ERBB2) or genomic rearrangements (e.g., ALK, ROS1, RET, NTRK) (1).
Many of these driver genetic alterations are currently targetable with specific therapy such as tyrosine kinase inhibitors (TKIs), that have significantly improved the prognosis of NSCLC.
Oncogene-addicted disease have distinctive clinical features; in some cases, different genetic aberrations are overlapping although some of these alterations are usually mutually exclusive.
Significant prognostic changes in recent years occurred for NSCLC oncogene addicted, even for uncommon oncogenic driver including ROS1 rearrangements. Next generation sequencing (NGS) has important role in ROS1-rearrangements detection and also for identification of resistance mutation to specific target agents, however this approach is not yet routinely available in clinical practice. The goal for the near future is detection of novel selective drugs for uncommon oncogene drivers and their optimal sequencing by evolving methods of molecular diagnostics.
This narrative review aims to outline the methods of ROS1 rearrangements detection and their potential evolution for the future. We also analysed the main therapeutic implications for NSCLC with ROS1 rearrangements. We present the following article in accordance with the Narrative Review reporting checklist (available at https://pcm.amegroups.com/article/view/10.21037/pcm-22-6/rc).
Methods
In this narrative review, we focused on detection pattern of ROS1 rearrangements and the consequent therapeutic implication in lung cancer. To this aim, we have analysed all the relevant literature in PubMed from 2007 to 2021 on evolving techniques for the molecular detection of ROS1 rearrangements and on the main mechanisms of resistance with consequent developments of more selective drugs (Table 1). The PubMed database was searched principally using the keywords “Non-small cell lung cancer”, “ROS1 rearrangements”. We excluded articles not published in English.
Table 1
| Item | Specification |
|---|---|
| Date of search | December 1, 2021 |
| Database and other sources searched | PubMed |
| Search terms used | ROS1-positive NSCLC |
| Timeframe | 2007–2021 |
| Inclusion and exclusion criteria | We only included studies published in English |
| Selection process | The selection process was conducted by the authors |
| Any additional considerations, if applicable | We included data from phase I/II studies |
ROS1, ROS proto-oncogene 1 receptor tyrosine kinase; NSCLC, non-small cell lung cancer.
Research was performed at both preclinical and clinical levels. For the preclinical part, we selected more than 50 publications on the implications of in situ laboratory and molecular biology techniques comparing these approaches; for the clinical part we collected the main publications on ROS1 rearranged oncogene-addicted NSCLCs and reported international guidelines. We included data from ten phase I/II trials testing efficacy and safety of TKIs targeting ROS1 rearrangement.
Biology, clinical and pathological features of ROS1 rearrangement
ROS1 gene is located on the short arm of chromosome 6 (6q22) and included in the subfamily of insulin tyrosine kinase receptors. Physiological role of ROS1 is still unclear, it is known to have homology with anaplastic lymphoma kinase protein (ALK) greater than 80% in the ATP binding site and kinase domains, but function and expression in adult tissue are uncertain (2). Genomic alterations of ROS1 lead to fusion with different gene partners (e.g., CD74, CCDC6, EZR, FIG, KDELR2, LRIG3, MSN, SDC4, SLC34A2, TMP3, TPD52L1) encoding for oncogenic driver proteins; the fusion usually results in constitutive activation of ROS1 receptor tyrosine kinase and consequently upregulation of downstream signaling pathways (JAK/STAT, PI3K/AKT and MAPK/ERK) involved in the control of cell proliferation (1-6).
ROS1 fusions are implicated in vivo and in vitro in the pathogenesis of several tumors; notably, such fusions were described for the first time in glioblastoma and subsequently observed in other tumors including NSCLC, angiosarcoma, colon and gastric carcinoma and ovarian cancers (3,7-12).
In NSCLCs, ROS1 rearrangements occur in 1–2% of cases, and are generally related to some clinical-histological features as adenocarcinoma histology with psammomatous calcification and solid growth with mucinous/cribriform patterns. Patients with ROS1 positive NSCLC, are often young and non-smokers or low smokers; brain metastases are present in approximately 40% of cases at diagnosis. With regards to molecular setting, ROS1-positive NSCLCs are usually wild-type for EGFR mutations, ALK rearrangements and other driver mutations, the rare cases of overlap between ROS1 rearrangements and other driver molecular alterations should be assessed by further analysis with molecular testing, and usually are not confirmed (13-18). Moreover, in recent years more rigorous genetic analyses of complete molecular profile identified the coexistence of ROS1 and other driver alterations in NSCLC, such as RET rearrangements and MET amplification, despite their low frequency, with important clinical and therapeutic implications (19-22).
ROS1 detection methods
Currently, a number of methods are used to detect ROS1 rearrangements, which vary depending on the molecule being studied such as protein [immunohistochemistry (IHC)], DNA [fluorescence in situ hybridization (FISH); next-generation sequencing (NGS)], and RNA [reverse transcription polymerase chain reaction (RT-PCR); NGS] (23,24). Generally, in situ analysis methods such as FISH and IHC, are routinely used in clinical practice. FISH is traditionally considered the gold standard for ROS1 and ALK rearrangement identification in lung cancer, using the same criteria; this technique is moderately expensive, laborious and inclined to false negative results. IHC is an efficient tool for the selection of ROS1-positive lung cancer, with higher sensitivity (over 90%) as compared to FISH, which is instead characterized by high specificity (more than 90%), but low specificity (less than 60%). Furthermore, IHC needs low operator requirements and has a shorter turnaround time for results, but it reveals to be less specific than FISH. Therefore, detection of ROS1-negative NSCLC (whether non-smokers or smokers with non-squamous NSCLC) by IHC, may not require a validation by FISH, thus reducing testing costs (21,22,24-38). On the other hand, FISH is traditionally considered the gold standard for the validation of ROS1 rearrangements in lung cancer. IHC is generally preferred to FISH on formalin-fixed and paraffin-embedded (FFPE) tissue biopsy, as well as on few tumor cells obtained from pleural or pericardial effusion (39,40). Conversely, FISH can be performed on both tissue and cytological samples and it is superior compared to IHC on non-bloody cytological swipes. Although FISH is characterized by higher specificity (more than 90%) compared to IHC, this method has the limit to be somewhat expensive, laborious and inclined to false negative results and it is for these reasons that molecular analyses by RT-PCR and NGS are additionally executed (39,40).
ROS1 rearrangements diagnosis in FISH
Two main patterns of ROS1 rearrangements can be detected by FISH: “break-apart” pattern and “atypical pattern” using one isolated fusion signal. The “break-apart” FISH pattern, that uses dual color probes, is the conventional approach for ROS1 identification. In presence of a rearrangements in tumor cells, two ends of ROS1 gene are spliced and the part containing the tyrosine kinase domain is fused with another partner to create a ROS1 fusion gene. This technique involves two probes that lap with a green fluorochrome the 3' (centromeric) part of the fusion breakpoint and with an orange fluorochrome the 5' (telomeric) part; the green fluorochrome results by the kinase-fusion domain of ROS1 gene (typical rearranged pattern). In the atypical pattern the single green signal (5' end) does not show a corresponding orange signal (3' end) in conjunction to a fused and/or split signals (isolated 3' ROS1 signals). To detect ROS1 fusion gene positivity the signal of rearrangements recorded in FISH has to reach a cut-off of 15% or more among 50 neoplastic cells (24,38,40,41). Some “break-apart” FISH assay for ROS1 fusion have the limit to fail in detecting intrachromosomal deletions which is emerging as an acquired resistance mechanism to osimertinib (EGFR inhibitor) in lung adenocarcinoma (42,43).
ROS1 rearrangements diagnosis in IHC
A consensus of experts reports that ROS1 IHC can be used as a screening test in advanced stage lung adenocarcinoma patients but that positive ROS1 IHC results should be confirmed by FISH or other molecular methods (44).
IHC analysis for ROS1 rearrangements can be performed by different amplification kits and detection systems and the monoclonal antibody D4D6 (cell signalling technology) is generally used. A relatively recent multicenter study provided real-world data on ROS1 rearrangements in patients with NSCLC, demonstrating that the new SP384 ROS1 IHC clone (Ventana medical systems) showed excellent sensitivity. Although to date, no FDA-approved IHC assay is advised for clinical routine, the two available antibodies (i.e., D4D6 and SP384) have demonstrated high performance in most studies (45).
Two different score systems have been developed to define tumor ROS1-positivity. The first one is based on IHC staining intensity according to a semi-quantitative scoring system as follows: (I) negative (score 0), (II) weak signal (1+), (III) moderate signal (2+), (IV) strong positive signal (3+). Tumor cells with ROS1-rearrangements have intense immunohistochemical staining, resulting 2+/3+ score. Another employed score system is the H-score, ranging from 0 to 300. This score is obtained by multiplying percentage of positive tumor cells and IHC staining intensity (from 0 to 3+), with positivity threshold for ROS1 rearrangements greater than 100 (24,26). At IHC, neoplastic cells with ROS1 rearrangements present finely granular cytoplasmic staining, that changes according to different fusions. An important feature in the ROS1 detection by IHC is the possibility to insert an external positive control; indeed, although ROS1 protein is absent in normal adult tissues, it can be detected in reactive alveolar type II pneumocytes and macrophages (24,41). Despite higher sensitivity but lower specificity of IHC compared to FISH, the two techniques correlate well when D4D6 clone is combined with high sensitivity amplification kits in IHC. For IHC diagnosis of ROS1 rearrangement there are other promising but less investigated monoclonal and polyclonal anti-ROS1 antibodies (24).
Molecular technologies: RT-PCR and NGS
An alternative method to validate IHC-based ROS1 positivity, is the RT-PCR, although this technique is at time less reliable when used alone, unless RNA-based anchored multiplex polymerase chain reaction library preparation followed by NGS is performed.
RT-PCR, starting from RNA, first converts RNA to complementary DNA (cDNA); then the cDNA is amplified to allow the detection of gene fusions. Despite high sensitivity, specificity and rapidity, the availability of good quality RNA samples may limit the application of RT-PCR for the detection of ROS1 rearrangements and prevent the identification of atypical ROS1 fusion patterns in routine practice. For these reasons, a combined assay involving IHC followed by multiplex RNA-based PCR and NGS was recently evaluated (24,46-49).
NGS is a technology that enables nucleic acid sequencing of multiple genes simultaneously and can be performed with both DNA and RNA. Although it is affected by the need for appropriate bioinformatic equipment and staff skilled in data testing and analysis (50-56), this technology has proved to be an essential tool in all areas of genomic and epigenomic basic and clinical research.
Indeed, NGS has high sensitivity and specificity, and can allow the identification of gene alterations with low sample amount from both FFPE tissue and circulating tumor DNA (ctDNA) in plasma. Notably, NGS can simultaneously detect known or novel ROS1 fusions, as well as rearrangements by analyzing short intron region of DNA (57-64).
However, RNA-based NGS is preferable to DNA-based NGS in that sequencing can be focused on coding sequences instead of introns (63). To note that, although high quality RNA is needed, particularly if FFPE samples are used, the RNA based NGS can also allow the detection of any fused genes or novel ones such as ROS1-GOPC, overcoming the limits of DNA-based NGS not covering intronic breakpoints (43).
Therefore, NGS provides many advantages and is a noteworthy tool in molecular diagnosis of lung cancer, even in analysis of small sample amounts that cannot be analyzed by traditional methods. Indeed, Interestingly, Lim et al. reported that 58% of 51 patients with wild-type lung cancer, diagnosed by standard molecular testing, had gene alterations as ROS1 and others (e.g., EGFR, MET, RET, ALK, BRAF, NTRK) found by using an NGS approach (57,62).
Another emerging technology for ROS1 rearrangements detection is represented by NanoString nCounter platform. Specifically, nCounter platform is able to detect multiple gene fusions simultaneously including ALK, RET and ROS1, using limited amounts of RNA. However, similarly to the previous RNA-based tools, very short or highly fragmented RNA samples (35).
Comparison of IHC, FISH, RT-PCR and NGS in detection of ROS1-rearrangements
However, to date, a number of approaches are used for the detection of ROS1 rearrangements, some drawbacks need to be considered.
In a retrospective study, a total of 107 patients with NSCLC, including 92 adenocarcinomas, 12 squamous-cell carcinomas and 2 adeno-squamous carcinomas, 11 samples resulted positive for ROS1 by IHC, only two of which were confirmed with NGS (36,65). In the same study, there was evidence of a high concordance in terms of sensitivity between RT-PCR and NGS in 12 samples resulted positive for ROS1 and ALK rearrangements (65). Similarly, Reguart et al. demonstrated a concordance of 87% and 86% in the evaluation of ROS1 between nCounter platform vs. IHC and FISH, respectively (35).
In several studies, ROS1 positivity rates in the same cohort of NSCLC patients were different when evaluated by multiple approaches other than IHC; ROS1 positivity in IHC has also been reported in non-neoplastic conditions, such as bronchial metaplasia, in reactive type II pneumocytes or macrophages, confirming that the sensitivity of this methodologies is higher than its specificity.
Therefore, the detection of ROS1 rearrangements in IHC should be confirmed by further analyses with different techniques such as FISH or NGS, although some disadvantages limit its routine application in the clinical practice. The main advantages and disadvantages of NGS and other technologies for ROS1 detection are summarized in Table 2 (22,24,33,35,65).
Table 2
| Technologies | Advantages | Drawbacks |
|---|---|---|
| IHC | Low cost; high sensitivity; low number of samples needed; easy to use | Antibody dependence; dependence on tissue fixation; dependence on subjective error in interpretation results |
| FISH | High reliability | Expensive; long and difficult procedure; high risk of false negative results |
| RT-PCR | High sensitivity; high specificity; low number of samples needed | Need to use FFPE samples with good quality; risk of error in the interpretation of the result |
| NGS | Several applications; available in research and in clinic practice; evolving high number of kits; limited time from sample preparation to results | Highly specialized equipment for data analysis; expensive in some countries; no possibility of standardization and/or application of standardized material in clinical practice |
ROS1, ROS proto-oncogene 1 receptor tyrosine kinase; IHC, immunohistochemistry; FISH, fluorescence in situ hybridization; RT-PCR, reverse-transcriptase polymerase chain reaction; NGS, next generation sequencing; FFPE, formalin-fixed and paraffin-embedded.
Targeted agents for ROS1-positive NSCLC
Several drugs belonging to the class of TKIs are effective in NSCLC with ROS1 rearrangement (Table 3); currently crizotinib, ceritinib and entrectinib are approved by the Food and Drug Administration (FDA), while only crizotinib and entrectinib have been approved by the European Medicines Agency (EMA), and their use achieved a significant impact in terms of efficacy for these patients. The most relevant clinical studies available to date have been summarized in Table 4.
Table 3
| TKIs | Molecular target |
|---|---|
| Crizotinib | ALK, ROS1, MET |
| Entrectinib | ROS1, NTRK, ALK, TRKA, TRKB, TRKC |
| Ceritinib | ALK, ROS1 |
| Lorlatinib | ALK, ROS1 |
| Brigatinib | ALK, ROS1 |
| Repotrectinib | ALK, ROS1 |
| Cabozantinib | VEGF, ROS1, RET, MER, KIT, TYRO3, TRKB, FLT3, TIE2 |
TKI, tyrosine kinase inhibitor; ROS1, ROS proto-oncogene 1 receptor tyrosine kinase.
Table 4
| Agent | Study | Patients (n) | Outcomes |
|---|---|---|---|
| Crizotinib | Shaw et al., 2019, PROFILE 1001 (66) (phase I) | 53 | ORR 72%, PFS 51.4 months (95% CI: 29.3 months–not reached) |
| Mazières et al., 2015, EUROS1 (67) (cohort retrospective trial) | 32 | DCR 87%, PFS 9.1 months | |
| Moro-Sibilot et al., 2018, Acsè trial (68) (phase II) | 37 | ORR 47.2%, PFS 5.5 months (95% CI: 4.6–9.1 months) | |
| Wu et al., 2018, Asian study (69) (phase II) | 127 | ORR 71.7%, PFS 15.8 months (95% CI: 12.9–24 months) | |
| Landi et al., 2019, METROS trial (70) (phase II) | 505 | ORR 65%, PFS 22.8 months (95% CI: 15.2–30.3 months) | |
| Entrectinib | Dziadziuszko et al., 2021 (71), ALKA-372-001, STARTRK-1, STARTRK-2 (pooled data from phase I/II studies) | 161 | ORR 67.1%, PFS 15.7 months (95% CI: 13.9–28.6 months); intracranial ORR 79.2%, intracranial PFS 12 months (95% CI: 6.2–19.3 months) |
| Cabozantinib | Sun et al., 2019 (72), case series | 4 | ORR 25%, PFS 4.9–13.8 months |
| Lorlatinib | Solomon et al., 2018 (73), phase II trial | 47 | ORR 26.5% (pre-treated patients) |
| Shaw et al., 2019 (74), phase I/II trial | 69 | ORR 62% (TKIs naïve patients), ORR 35% (pre-treated patients) | |
| Repotrectinib | Cho et al., 2019, TRIDENT-1 (75) (phase I) | 75 | ORR 39% (pre-treated patients) |
ROS1, ROS proto-oncogene 1 receptor tyrosine kinase; NSCLC, non-small cell lung cancer; ORR, objective response rate; DCR, disease control rate; PFS, progression free survival; TKIs, tyrosine kinase inhibitors.
There are also emerging preclinical efficacy data, in vivo in vitro, of new generations ROS1-inhibitors, ROS1 NUV-520 and DS-6051b, overcoming G2032R resistance mutations in kinase domain secondary to crizotinib therapy and also not accessible by others target agents as lorlatinib and entrectinib (76,77). Furthermore NUV-520 is a brain-penetrant molecule, with preliminary data on activity in the central nervous system (CNS) (77).
Crizotinib
Crizotinib is an orally available TKI that binds the tyrosine kinase domain and blocks ATP-dependent functions in tumoral cells, thus inhibiting cell proliferation activity induced by several oncogene alterations including ROS1 as well as ALK and MET (Figure 1). Crizotinib has represented for years the gold standard in first line treatment for ROS1-positive NSCLC, according to the results of several preclinical, phase I, retrospective and prospective studies (2,69,78,79). The first efficacy data derive from PROFILE 1001, a phase I trial that included 53 patients with ROS1-rearranged NSCLC treated with crizotinib; at 22.4 months of treatment the objective response rate (ORR) was 72%, of which six patients (11%) with complete response (63). In the updated trial results of 2019, 19.3 months (95% CI: 15.2–39.1 months) of median progression free survival (PFS) and 51.4 months (95% CI: 29.3 months–not reached) of median overall survival (OS) were reported; no correlation between crizotinib efficacy and ROS1 fusion partner was observed and a favorable safety profile was shown even with prolonged treatment (66). All patients had at least one adverse event (AEs), albeit mostly (n=33, 64%) of grade 1–2 according to Common Terminology Criteria for Adverse Events (CTCAE); only 19 patients (35%) had grade 3 AEs (neutropenia, hypophosphatemia, elevated transaminases) and no grade 4 events were reported. The most common side effects were vision disorder, nausea, edema, diarrhea; no AEs were related to permanent discontinuation of treatment (66).
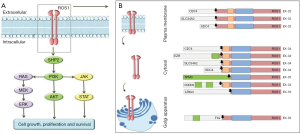
Following these results, efficacy of crizotinib in ROS1-positive, advanced NSCLC was demonstrated in other prospective and retrospective studies (67,69,79); in the EUROS1 cohort retrospective study, crizotinib demonstrated disease control rate (DCR) of 87% and PFS of 44% at 12 months in 32 patients with ROS1-positive NSCLC (77). Two subsequent prospective phase II trials confirmed the efficacy of crizotinib with a DCR higher than 85%, although reporting different PFS results, respectively 5.5 months (95% CI: 4.6–9.1 months) in the French study and 15.9 months (95% CI: 12.9–24.0 months) in the Asian study (69,79). The different PFS between these trials, particularly the relatively short result in the Acsè trial, may be influenced by the small size of the study population (i.e., 37 patients in the French study vs. 127 patients in the Asian trial) (67,69,79). In the METROS trial, a prospective phase II study, Landi et al. analyzed the clinical role of crizotinib in 505 patients with advanced NSCLC and ROS1 rearrangements or MET amplifications (68,70); in the ROS1-positive cohort, the ORR was 65%, including 4% complete responses, with DCR of 85% and a median PFS of 22.8 months (95% CI: 15.2–30.3 months). Overall, ROS1-positive population experienced treatment-related AE mostly of grade 1–2 and only 4% of patients reported serious events (grade 3–4) as neutropenia, peripheral edema fatigue, nausea (68,70).
In spite of these results, the efficacy of crizotinib is limited by different mechanisms of resistance, such as point mutations or activation of alternative signaling pathways, that consequently lead to loss of drug activity and disease progression. Several mutations in different domains were detected (e.g., D2033N, G2032R, L2026M, L2155S) in patients with advanced NSCLC that, through various paths, lead to an increase in kinase activity and therefore in cell proliferation. The G2032R mutation, involving the kinase domain target of the drug, is apparently the most common way of resistance (80-86).
In several studies, the activation of the EGFR signaling pathway was observed in advanced ROS1-positive NSCLC treated with crizotinib, leading to loss of ROS1 pathway cell proliferation signal and consequent resistance to treatment. While in some tumoral cells KIT signaling pathway resulted activated, highlighting the possibility of an association therapy with selective alternative pathway-inhibitors, such as erlotinib or gefitinib for EGFR and ponatinib for KIT, to overcome resistance to crizotinib in selected cases. The role of human epidermal growth factor receptor 2 (HER2) and TP53 gene amplification and mitogen-activated protein kinase (MAPK) pathway upregulation in crizotinib resistance remains still unclear (86-91).
Entrectinib
Entrectinib is another oral TKI currently available and effective for the treatment of ROS1-rearranged NSCLC, particularly active on brain metastases due to its potential to cross the blood-brain barrier (BBB). Although there are no trials comparing entrectinib and crizotinib, a retrospective analysis showed that ROS1-positive patients treated with crizotinib, without brain metastases at diagnosis, had an encephalic progression during treatment in 42% of cases with a time to progression of 24 months (13).
Whereas, recent results from integrate analysis of phase I/II studies on entrectinib as first-line in 161 patients with advanced NSCLC, have shown high clinical benefit this TKI with a follow-up over 6 months. The objective response was obtained and confirmed in 108 patients (ORR 67.1%) in term of overall efficacy, with a median duration of response (DoR) of 15.7 months. In 24 patients with brain metastases at diagnosis, the intracranial ORR was 79.2% with median PFS of 12.9 months (95% CI: 6.2–19.3 months) and median DoR of 12.9 months. Interestingly, the time to intracranial response was short (median: 0.95 months) (71,92-94). With regard to safety profile, patients treated with entrectinib reported mostly low grade and manageable AEs, such as dysgeusia or fatigue, usually reversible (71). Therefore, entrectinib showed benefit in patients with intracranial disease, and a good safety profile, similar to crizotinib and others TKIs; nevertheless, it has limited efficacy data against ROS1 resistance mutations like G2032R, D2033N and L2026M (92).
Other agents
Other TKIs acting against ROS1 alterations, including brigatinib, cabozantinib, ceritinib, lorlatinib and repotrectinib shown efficacy in ROS1 positive lung cancer, but with limited activity against resistance mutations of ROS1-kinase domain following first line therapy with crizotinib; furthermore, these drugs achieved brain disease control (83,95,96). While brigatinib, lorlatinib and ceritinib do not appear to be effective in crizotinib-pretreated NSCLC when G2032R resistance mutation develops, other agents including Repotrectinib, result to have modest activity in vitro and in vivo in this setting (97).
Cabozantinib is an orally available multi-target TKI (active on AXL, KIT, MET, RET, ROS1, VEGFR2, TIE2), currently available for renal cell and medullary thyroid carcinoma. Cabozantinib showed efficacy in ROS1-positive NSCLC patients who develop resistance to crizotinib and ceritinib; more specifically, Sun et al. reported the results from a case series of four previously treated patients (three of which had received both crizotinib and ceritinib); in this series, cabozantinib achieved a 100% DCR and OR was observed in one patient, while median PFS ranged from 4.9 to 13.8 months (72). However, although cabozantinib achieved promising results in overcoming acquired resistance ROS1 mutations, this agent is characterized by a challenging toxicity profile compared to other TKIs, with the most common reported AE being neutropenia, xeroderma and pulmonary embolism (71,82).
Lorlatinib and repotrectinib, which are potent oral inhibitors of ALK and ROS1, among other targets, achieved promising results against several ROS1 emerging mutations (D2033N e S1986Y) resistant to crizotinib and ceritinib, but not against G2032R, common resistance mutations after previous treatment with crizotinib. In phase I/II studies, lorlatinib and repotrectinib demonstrated ORR of 26.5% and 39%, respectively, in patients pretreated with crizotinib or other TKIs; furthermore, both agents achieved important benefit for the management of intracranial disease, due to high potential to cross the BBB, with response rate with repotrectinib close to 100% in treatment-naïve patients and 75% in pretreated patients. Finally, both lorlatinib and repotrectinib are characterized by a globally manageable safety profile and high activity in ROS1-positive disease especially on CNS disease. While the use of repotrectinib in clinical practice is still experimental, lorlatinib currently represents the preferred second-line treatment for ROS1-positive, pretreated NSCLC, due to its ability to overcome acquired resistance (72-75,85,98-104).
Discussion
The treatment of NSCLC has undergone profound changes in recent years. The definition of “wild type” NSCLC is changing from year to year thanks to the increase of drugs active towards specific genic alterations, including known and novel oncogenic drivers; as a result, antineoplastic therapy is being increasingly tailored to each individual patient with important and prolonged benefits in terms of response and survival. In this setting, the availability of new drugs must be accompanied by a cultural change. To date, despite the consolidated availability of targeted agents, many patients are still not tested for the four most frequent molecular targets, which include EGFR, ALK, BRAF and ROS1. Quite surprisingly, only approximately 50% of patients are being tested in US for all the aforementioned targets, in addition to PD-L1 expression. ROS1 is tested in approximately 70–75% of cases (105,106). This change of perspective must be accompanied by an improvement in the knowledge of oncologists, pathologists and molecular biologists who research these alterations. As previously discussed, the appropriate identification of ROS1 rearrangement can present some diagnostic challenges depending on the method used. NGS is not widely available yet, especially in peripheric area hospitals, and other methods have non-negligible limits in terms of sensitivity and specificity.
Among the advantages of NGS approach, one specific mention should be reserved to its ability to detect acquired resistance mutations which develop during targeted treatment; while this approach is useful for virtually all cases of oncogene-addicted NSCLC, including novel emerging biomarkers (107), it appears even more appealing when ROS1 or ALK rearrangements are involved, as the identification of specific on-target resistance mutation might help select the most appropriate compound among the different available agents based on their individual ability to overcome that specific mechanism. Furthermore, NGS approaches can be employed on peripheral blood, eventually leading to decreased need of invasive tissue biopsies (108).
Notably, the increasing availability of novel effective targeted agents for ROS1-positive NSCLC may lead to two opposing strategies: one possible approach involves the sequential use of available agents based on the identified emerging mutations, while the other approach relies on the upfront use of the most active agents. While the former approach appears to be more tailored on each individual patient and provides a strong rationale for therapeutic sequences, thus offering credible therapeutic opportunities after the initial disease progression, the latter approach is based on preventing the development of resistance mechanisms in first place and implies reduced need for repeated molecular analyses throughout each patient’s history. Currently, the “best first” approach is being favored, at least partially due to its easy application compared to sequential strategies; furthermore, the activity of novel agents on preventing and treating brain metastases, compared to crizotinib, cannot be ignored, as it can translate in relevant prevention or improvement of neurologic symptoms. In contrast with other more frequently detected oncogenic drivers, such as EGFR, results from randomized controlled trials comparing different agents for ROS1-positive NSCLC are currently lacking; hence, most available evidence to date derives from single-arm phase I/II trials. While randomized trials are generally warranted, the rarity of ROS1 rearrangements implies that such trials suffer from slow enrollment, which might make their results obsolete, as additional novel agents are in the meanwhile developed. In this setting, real-life data collected with coordinated, multi-institutional efforts, might provide useful data; furthermore, in the near future, the investigation of uncommon oncogenic drivers for NSCLC, for which ROS1 represents a good model, will require novel methodic designs, in order to provide robust and useful data while at the same time keeping pace with the rapid development of novel agents and sequencing technologies.
Acknowledgments
Funding: This work was supported by the Italian Ministry of Health (5x1000 funds; No. CO-2016-02361470; Ricerca Corrente 2022-2024) and the Bristol-Myers-Squibb (No. BMS 209-828) for grants in support of research on non-small cell lung cancer.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Fabrizio Tabbò, Umberto Malapelle, Maria Lucia Reale and Angela Listì) for the series “How to Detect and Treat NSCLC Patients with Oncogenic Fusions” published in Precision Cancer Medicine. The article has undergone external peer review.
Reporting Checklist: The authors have completed the Narrative Review reporting checklist. Available at https://pcm.amegroups.com/article/view/10.21037/pcm-22-6/rc
Peer Review File: Available at https://pcm.amegroups.com/article/view/10.21037/pcm-22-6/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://pcm.amegroups.com/article/view/10.21037/pcm-22-6/coif). The series “How to Detect and Treat NSCLC Patients with Oncogenic Fusions” was commissioned by the editorial office without any funding or sponsorship. GR received honoraria for Novartis, BMS and Roche. MT received travel grants by Roche, BMS, Astrazeneca and Takeda; received honoraria as medical writer for Novartis and Amgen. GB decalred the consulting fees by Boeringher Inghelaim and Astrazeneca; received honoraria for Bristol, Astrazeneca and Roche; received travel grants by Roche, Astrazeneca and Pierre Fabre; served on Advisory Board established by Boeringher, Astrazeneca and Pierre Fabre. CD received honoraria for Astrazeneca, BMS and Roche. EB received honoraria for BMS; received travel grants by BMS, MSD. ETT received honoraria for Merck Sharp and Dohme and BMS. ER received honoraria for BMS, MSA, Astrazeneca and Roche; served on an Advisory Board established by SANOFI. CG received honoraria from Astrazeneca, BMS and Roche. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Takeuchi K, Soda M, Togashi Y, et al. RET, ROS1 and ALK fusions in lung cancer. Nat Med 2012;18:378-81. [Crossref] [PubMed]
- Roskoski R Jr. ROS1 protein-tyrosine kinase inhibitors in the treatment of ROS1 fusion protein-driven non-small cell lung cancers. Pharmacol Res 2017;121:202-12. [Crossref] [PubMed]
- Rikova K, Guo A, Zeng Q, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell 2007;131:1190-203. [Crossref] [PubMed]
- Gainor JF, Shaw AT. Novel targets in non-small cell lung cancer: ROS1 and RET fusions. Oncologist 2013;18:865-75. [Crossref] [PubMed]
- Lin JJ, Shaw AT. Recent Advances in Targeting ROS1 in Lung Cancer. J Thorac Oncol 2017;12:1611-25. [Crossref] [PubMed]
- Zhang Y, Yu M, Yuan M, et al. Identification of a Novel RBPMS-ROS1 Fusion in an Adolescent Patient With Microsatellite-instable Advanced Lung Adenocarcinoma Sensitive to Crizotinib: A Case Report. Clin Lung Cancer 2020;21:e78-83. [Crossref] [PubMed]
- Birchmeier C, Sharma S, Wigler M. Expression and rearrangement of the ROS1 gene in human glioblastoma cells. Proc Natl Acad Sci U S A 1987;84:9270-4. [Crossref] [PubMed]
- Gu TL, Deng X, Huang F, et al. Survey of tyrosine kinase signaling reveals ROS kinase fusions in human cholangiocarcinoma. PLoS One 2011;6:e15640. [Crossref] [PubMed]
- Giacomini CP, Sun S, Varma S, et al. Breakpoint analysis of transcriptional and genomic profiles uncovers novel gene fusions spanning multiple human cancer types. PLoS Genet 2013;9:e1003464. [Crossref] [PubMed]
- Birch AH, Arcand SL, Oros KK, et al. Chromosome 3 anomalies investigated by genome wide SNP analysis of benign, low malignant potential and low grade ovarian serous tumours. PLoS One 2011;6:e28250. [Crossref] [PubMed]
- Lee J, Lee SE, Kang SY, et al. Identification of ROS1 rearrangement in gastric adenocarcinoma. Cancer 2013;119:1627-35. [Crossref] [PubMed]
- Aisner DL, Nguyen TT, Paskulin DD, et al. ROS1 and ALK fusions in colorectal cancer, with evidence of intratumoral heterogeneity for molecular drivers. Mol Cancer Res 2014;12:111-8. [Crossref] [PubMed]
- Patil T, Smith DE, Bunn PA, et al. The Incidence of Brain Metastases in Stage IV ROS1-Rearranged Non-Small Cell Lung Cancer and Rate of Central Nervous System Progression on Crizotinib. J Thorac Oncol 2018;13:1717-26. [Crossref] [PubMed]
- Bergethon K, Shaw AT, Ou SH, et al. ROS1 rearrangements define a unique molecular class of lung cancers. J Clin Oncol 2012;30:863-70. [Crossref] [PubMed]
- Lin JJ, Ritterhouse LL, Ali SM, et al. ROS1 Fusions Rarely Overlap with Other Oncogenic Drivers in Non-Small Cell Lung Cancer. J Thorac Oncol 2017;12:872-7. [Crossref] [PubMed]
- Facchinetti F, Rossi G, Bria E, et al. Oncogene addiction in non-small cell lung cancer: Focus on ROS1 inhibition. Cancer Treat Rev 2017;55:83-95. [Crossref] [PubMed]
- Lin JJ, Langenbucher A, Gupta P, et al. Small cell transformation of ROS1 fusion-positive lung cancer resistant to ROS1 inhibition. NPJ Precis Oncol 2020;4:21. [Crossref] [PubMed]
- Wiesweg M, Eberhardt WEE, Reis H, et al. High Prevalence of Concomitant Oncogene Mutations in Prospectively Identified Patients with ROS1-Positive Metastatic Lung Cancer. J Thorac Oncol 2017;12:54-64. [Crossref] [PubMed]
- Tang Z, Zhang J, Lu X, et al. Coexistent genetic alterations involving ALK, RET, ROS1 or MET in 15 cases of lung adenocarcinoma. Mod Pathol 2018;31:307-12. [Crossref] [PubMed]
- Mao Y, Wu S. ALK and ROS1 concurrent with EGFR mutation in patients with lung adenocarcinoma. Onco Targets Ther 2017;10:3399-404. [Crossref] [PubMed]
- Davies KD, Le AT, Theodoro MF, et al. Identifying and targeting ROS1 gene fusions in non-small cell lung cancer. Clin Cancer Res 2012;18:4570-9. [Crossref] [PubMed]
- Sholl LM, Sun H, Butaney M, et al. ROS1 immunohistochemistry for detection of ROS1-rearranged lung adenocarcinomas. Am J Surg Pathol 2013;37:1441-9. [Crossref] [PubMed]
- Lindeman NI, Cagle PT, Aisner DL, et al. Updated Molecular Testing Guideline for the Selection of Lung Cancer Patients for Treatment With Targeted Tyrosine Kinase Inhibitors: Guideline From the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. Arch Pathol Lab Med 2018;142:321-46. [Crossref] [PubMed]
- Rossi G, Jocollé G, Conti A, et al. Detection of ROS1 rearrangement in non-small cell lung cancer: current and future perspectives. Lung Cancer (Auckl) 2017;8:45-55. [Crossref] [PubMed]
- Suehara Y, Arcila M, Wang L, et al. Identification of KIF5B-RET and GOPC-ROS1 fusions in lung adenocarcinomas through a comprehensive mRNA-based screen for tyrosine kinase fusions. Clin Cancer Res 2012;18:6599-608. [Crossref] [PubMed]
- Viola P, Maurya M, Croud J, et al. A Validation Study for the Use of ROS1 Immunohistochemical Staining in Screening for ROS1 Transloca- tions in Lung Cancer. J Thorac Oncol 2016;11:1029-39. [Crossref] [PubMed]
- Yoshida A, Tsuta K, Wakai S, et al. Immunohistochemical detection of ROS1 is useful for identifying ROS1 rearrangements in lung cancers. Mod Pathol 2014;27:711-20. [Crossref] [PubMed]
- Mescam-Mancini L, Lantuejoul S, Moro-Sibilot D, et al. On the rel- evance of a testing algorithm for the detection of ROS1-rearranged lung adenocarcinomas. Lung Cancer 2014;83:168-73. [Crossref] [PubMed]
- Shan L, Lian F, Guo L, et al. Detection of ROS1 gene rearrangement in lung adenocarcinoma: comparison of IHC, FISH and real-time RT-PCR. PLoS One 2015;10:e0120422. [Crossref] [PubMed]
- Boyle TA, Masago K, Ellison KE, et al. ROS1 immuno- histochemistry among major genotypes of non-small-cell lung cancer. Clin Lung Cancer 2015;16:106-11. [Crossref] [PubMed]
- Cao B, Wei P, Liu Z, et al. Detection of lung adenocarcinoma with ROS1 rearrangement by IHC, FISH, and RT-PCR and analysis of its clinicopathologic features. Onco Targets Ther 2015;9:131-8. [PubMed]
- Rogers TM, Russell PA, Wright G, et al. Comparison of methods in the detection of ALK and ROS1 rearrangements in lung cancer. J Thorac Oncol 2015;10:611-8. [Crossref] [PubMed]
- Cha YJ, Lee JS, Kim HR, et al. Screening of ROS1 rearrangements in lung adenocarcinoma by immunohistochemistry and comparison with ALK rearrangements. PLoS One 2014;9:e103333. [Crossref] [PubMed]
- Selinger CI, Li BT, Pavlakis N, et al. Screening for ROS1 gene rear- rangements in non-small-cell lung cancers using immunohistochemistry with FISH con rmation is an effective method to identify this rare target. Histopathology 2017;70:402-11. [Crossref] [PubMed]
- Reguart N, Teixidó C, Giménez-Capitán A, et al. Identification of ALK, ROS1, and RET fusions by a multiplexed mRNA-based assay in formalin- xed, paraf n-embedded samples from advanced non-small- cell lung cancer patients. Clin Chem 2017;63:751-60. [Crossref] [PubMed]
- Su Y, Goncalves T, Dias-Santagata D, et al. Immunohisto- chemical Detection of ROS1 Fusion. Am J Clin Pathol 2017;147:77-82. [PubMed]
- Warth A, Muley T, Dienemann H, et al. ROS1 expression and translo- cations in non-small-cell lung cancer: clinicopathological analysis of 1478 cases. Histopathology 2014;65:187-94. [Crossref] [PubMed]
- Bozzetti C, Nizzoli R, Tiseo M, et al. ALK and ROS1 rearrangements tested by fluorescence in situ hybridization in cytological smears from advanced non-small cell lung cancer patients. Diagn Cytopathol 2015;43:941-6. [Crossref] [PubMed]
- Jurmeister P, Lenze D, Berg E, et al. Parallel screening for ALK, MET and ROS1 alterations in non-small cell lung cancer with implications for daily routine testing. Lung Cancer 2015;87:122-9. [Crossref] [PubMed]
- Leary RJ, Kinde I, Diehl F, et al. Development of personalized tumor biomarkers using massively parallel sequencing. Sci Transl Med 2010;2:20ra14. [Crossref] [PubMed]
- Bubendorf L, Büttner R, Al-Dayel F, et al. Testing for ROS1 in non-small cell lung cancer: a review with recommendations. Virchows Arch 2016;469:489-503. [Crossref] [PubMed]
- Savic S, Bubendorf L. Role of fluorescence in situ hybridization in lung cancer cytology. Acta Cytol 2012;56:611-21. [Crossref] [PubMed]
- Zeng L, Yang N. GOPC-ROS1 Rearrangements as an Acquired Resistance Mechanism to Osimertinib and Responding to Crizotinib Combined Treatments in Lung Adenocarcinoma. J Thorac Oncol 2018;13:e114-6. [Crossref] [PubMed]
- Garrido P, Conde E, de Castro J, et al. Updated guidelines for predictive biomarker testing in advanced non-small-cell lung cancer: a National Consensus of the Spanish Society of Pathology and the Spanish Society of Medical Oncology. Clin Transl Oncol 2020;22:989-1003. [Crossref] [PubMed]
- Conde E, Hernandez S, Martinez R, et al. Assessment of a New ROS1 Immunohistochemistry Clone (SP384) for the Identification of ROS1 Rearrangementss in Patients with Non-Small Cell Lung Carcinoma: the ROSING Study. J Thorac Oncol 2019;14:2120-32. [Crossref] [PubMed]
- Rossi G, Ragazzi M, Tamagnini I, et al. Does immunohistochemistry represent a robust alternative technique in determining drugable predictive gene alterations in non-small cell lung cancer? Curr Drug Targets 2017;18:13-26. [Crossref] [PubMed]
- Lira ME, Choi YL, Lim SM, et al. A single-tube multiplexed assay for detecting ALK, ROS1, and RET fusions in lung cancer. J Mol Diagn 2014;16:229-43. [Crossref] [PubMed]
- Wong SQ, Li J, Tan AY, et al. Sequence artefacts in a prospective series of formalin-fixed tumours tested for mutations in hotspot regions by massively parallel sequencing. BMC Med Genomics 2014;7:23. [Crossref] [PubMed]
- Zheng Z, Liebers M, Zhelyazkova B, et al. Anchored multiplex PCR for targeted next-generation sequencing. Nat Med 2014;20:1479-84. [Crossref] [PubMed]
- Cirulli ET, Goldstein DB. Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nat Rev Genet 2010;11:415-25. [Crossref] [PubMed]
- Sultan M, Schulz MH, Richard H, et al. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science 2008;321:956-60. [Crossref] [PubMed]
- Ju YS, Kim JI, Kim S, et al. Extensive genomic and transcriptional diversity identified through massively parallel DNA and RNA sequencing of eighteen Korean individuals. Nat Genet 2011;43:745-52. [Crossref] [PubMed]
- Behjati S, Tarpey PS. What is next generation sequencing? Arch Dis Child Educ Pract Ed 2013;98:236-8. [Crossref] [PubMed]
- Levy SE, Myers RM. Advancements in Next-Generation Sequencing. Annu Rev Genomics Hum Genet 2016;17:95-115. [Crossref] [PubMed]
- Slatko BE, Gardner AF, Ausubel FM. Overview of Next-Generation Sequencing Technologies. Curr Protoc Mol Biol 2018;122:e59. [Crossref] [PubMed]
- Pereira MA, Malta FSV, Freire MCM, et al. Application of Next-Generation Sequencing in the Era of Precision Medicine. In Applications of RNA-Seq and Omics Strategies—From Microorganisms to Human Health. IntechOpen: London, UK: 2017.
- Lim SM, Kim EY, Kim HR, et al. Genomic profiling of lung adenocarcinoma patients reveals therapeutic targets and confers clinical benefit when standard molecular testing is negative. Oncotarget 2016;7:24172-8. [Crossref] [PubMed]
- Taylor C, Chacko S, Davey M, et al. Peptide-Affinity Precipitation of Extracellular Vesicles and Cell-Free DNA Improves Sequencing Performance for the Detection of Pathogenic Mutations in Lung Cancer Patient Plasma. Int J Mol Sci 2020;21:9083. [Crossref] [PubMed]
- Govind KB, Koppaka D, Dasappa L, et al. Detection of clinically relevant epidermal growth factor receptor pathway mutations in circulating cell-free tumor DNA using next generation sequencing in squamous cell carcinoma lung. South Asian J Cancer 2019;8:247-9. [Crossref] [PubMed]
- Yamamoto G, Kikuchi M, Kobayashi S, et al. Routine genetic testing of lung cancer specimens derived from surgery, bronchoscopy and fluid aspiration by next generation sequencing. Int J Oncol 2017;50:1579-89. [Crossref] [PubMed]
- Li H, Xie Y, Lin Y, et al. Different Gene Mutation Spectrum of the Paired CSF and Plasma Samples in Lung Adenocarcinoma with Leptomeningeal Metastases: The Liquid Biopsy Based on Circulating Tumor DNA. Zhongguo Fei Ai Za Zhi 2020;23:646-54. [PubMed]
- Cainap C, Balacescu O, Cainap SS, et al. Next Generation Sequencing Technology in Lung Cancer Diagnosis. Biology (Basel) 2021;10:864. [Crossref] [PubMed]
- Davies KD, Le AT, Sheren J, et al. Comparison of Molecular Testing Modalities for Detection of ROS1 Rearrangements in a Cohort of Positive Patient Samples. J Thorac Oncol 2018;13:1474-82. [Crossref] [PubMed]
- Benayed R, Offin M, Mullaney K, et al. High Yield of RNA Sequencing for Targetable Kinase Fusions in Lung Adenocarcinomas with No Mitogenic Driver Alteration Detected by DNA Sequencing and Low Tumor Mutation Burden. Clin Cancer Res 2019;25:4712-22. [Crossref] [PubMed]
- Nong L, Zhang Z, Xiong Y, et al. Comparison of next-generation sequencing and immunohistochemistry analysis for targeted therapy-related genomic status in lung cancer patients. J Thorac Dis 2019;11:4992-5003. [Crossref] [PubMed]
- Shaw AT, Riely GJ, Bang YJ, et al. Crizotinib in ROS1-rearranged advanced non-small-cell lung cancer (NSCLC): updated results, including overall survival, from PROFILE 1001. Ann Oncol 2019;30:1121-6. [Crossref] [PubMed]
- Mazières J, Zalcman G, Crinò L, et al. Crizotinib therapy for advanced lung adenocarcinoma and a ROS1 rearrangement: results from the EUROS1 cohort. J Clin Oncol 2015;33:992-9. [Crossref] [PubMed]
- Moro-Sibilot D, Cozic N. OA12.03 activity of crizotinib in MET or ROS1 positive (þ) NSCLC: results of the AcSè Trial. J Thorac Oncol 2018;13:S348. [Crossref]
- Wu YL, Yang JC, Kim DW, et al. Phase II Study of Crizotinib in East Asian Patients With ROS1-Positive Advanced Non-Small-Cell Lung Cancer. J Clin Oncol 2018;36:1405-11. [Crossref] [PubMed]
- Landi L, Chiari R, Tiseo M, et al. Crizotinib in MET-Deregulated or ROS1-Rearranged Pretreated Non-Small Cell Lung Cancer (METROS): A Phase II, Prospective, Multicenter, Two-Arms Trial. Clin Cancer Res 2019;25:7312-9. [Crossref] [PubMed]
- Dziadziuszko R, Krebs MG, De Braud F, et al. Update Integrated Analysis of the Efficacy and Safety of Entrectinib in locally Advanced or Metastatic ROS1Fusion-Positive Non-Small-Cell-Lung Cancer. J Clin Oncol 2021;39:1253-63. [Crossref] [PubMed]
- Sun TY, Niu X, Chakraborty A, et al. Lengthy Progression-Free Survival and Intracranial Activity of Cabozantinib in Patients with Crizotinib and Ceritinib-Resistant ROS1-Positive Non-Small Cell Lung Cancer. J Thorac Oncol 2019;14:e21-4. [Crossref] [PubMed]
- Solomon BJ, Besse B, Bauer TM, et al. Lorlatinib in patients with ALK-positive non-small-cell lung cancer: results from a global phase 2 study. Lancet Oncol 2018;19:1654-67. [Crossref] [PubMed]
- Shaw AT, Solomon BJ, Chiari R, et al. Lorlatinib in advanced ROS1-positive non-small-cell lung cancer: a multicentre, open-label, single-arm, phase 1-2 trial. Lancet Oncol 2019;20:1691-701. [Crossref] [PubMed]
- Cho BC, Drilon AE, Doebele RC, et al. Safety and Preliminary Clinical Activity of Repotrectinib in Patients with Advanced ROS1 Fusion-Positive Non-Small Cell Lung Cancer (TRIDENT-1 Study). J Clin Oncol 2019;37:9011. [Crossref]
- Katayama R, Gong B, Togashi N, et al. The new-generation selective ROS1/NTRK inhibitor DS-6051b overcomes crizotinib resistant ROS1-G2032R mutation in preclinical models. Nat Commun 2019;10:3604. [Crossref] [PubMed]
- Pelish HE. Abstract 1465: NUV-520 (NVL-520) is a brain-penetrant and highly selective ROS1 inhibitors with antitumor activity against the G2032R solvent front mutation. Cancer Res 2021;81:1465. [Crossref]
- Shaw AT, Ou SH, Bang YJ, et al. Crizotinib in ROS1-rearranged non-small-cell lung cancer. N Engl J Med 2014;371:1963-71. [Crossref] [PubMed]
- Moro-Sibilot D, Cozic N, Pérol M, et al. Crizotinib in c-MET- or ROS1-positive NSCLC: results of the AcSé phase II trial. Ann Oncol 2019;30:1985-91. [Crossref] [PubMed]
- McCoach CE, Le AT, Gowan K, et al. Resistance Mechanisms to Targeted Therapies in ROS1þ and ALKþ Non–Small Cell Lung Cancer. Clin Cancer Res 2018;24:3334-47. [Crossref] [PubMed]
- Facchinetti F, Loriot Y, Kuo MS, et al. Crizotinib-Resistant ROS1 Mutations Reveal a Predictive Kinase Inhibitor Sensitivity Model for ROS1- and ALK-Rearranged Lung Cancers. Clin Cancer Res 2016;22:5983-91. [Crossref] [PubMed]
- Drilon A, Somwar R, Wagner JP, et al. A Novel Crizotinib-Resistant Solvent-Front Mutation Responsive to Cabozantinib Therapy in a Patient with ROS1-Rearranged Lung Cancer. Clin Cancer Res 2016;22:2351-8. [Crossref] [PubMed]
- Awad MM, Katayama R, McTigue M, et al. Acquired resistance to crizotinib from a mutation in CD74-ROS1. N Engl J Med 2013;368:2395-401. [Crossref] [PubMed]
- Gainor JF, Friboulet L, Yoda S, et al. Frequency and Spectrum of ROS1 Resistance Mutations in ROS1-Positive Lung Cancer Patients Progressing on Crizotinib. J Clin Oncol 2016;34:9072. [Crossref]
- Zou HY, Li Q, Engstrom LD, et al. PF-06463922 is a potent and selective next-generation ROS1/ALK inhibitor capable of blocking crizotinib-resistant ROS1 mutations. Proc Natl Acad Sci U S A 2015;112:3493-8. [Crossref] [PubMed]
- Song A, Kim TM, Kim DW, et al. Molecular Changes Associated with Acquired Resistance to Crizotinib in ROS1-Rearranged Non-Small Cell Lung Cancer. Clin Cancer Res 2015;21:2379-87. [Crossref] [PubMed]
- Dziadziuszko R, Le AT, Wrona A, et al. An Activating KIT Mutation Induces Crizotinib Resistance in ROS1-Positive Lung Cancer. J Thorac Oncol 2016;11:1273-81. [Crossref] [PubMed]
- Dagogo-Jack I, Rooney M, Nagy RJ, et al. Molecular Analysis of Plasma From Patients With ROS1-Positive NSCLC. J Thorac Oncol 2019;14:816-24. [Crossref] [PubMed]
- Katayama R, Shaw AT, Khan TM, et al. Mechanisms of acquired crizotinib resistance in ALK-rearranged lung Cancers. Sci Transl Med 2012;4:120ra17. [Crossref] [PubMed]
- Sasaki T, Koivunen J, Ogino A, et al. A novel ALK secondary mutation and EGFR signaling cause resistance to ALK kinase inhibitors. Cancer Res 2011;71:6051-60. [Crossref] [PubMed]
- Doebele RC, Pilling AB, Aisner DL, et al. Mechanisms of resistance to crizotinib in patients with ALK gene rearranged non-small cell lung cancer. Clin Cancer Res 2012;18:1472-82. [Crossref] [PubMed]
- Facchinetti F, Friboulet L. Profile of entrectinib and its potential in the treatment of ROS1-positive NSCLC: evidence to date. Lung Cancer (Auckl) 2019;10:87-94. [Crossref] [PubMed]
- Drilon A, Siena S, Ou SI, et al. Safety and Antitumor Activity of the Multitargeted Pan-TRK, ROS1, and ALK Inhibitor Entrectinib: Combined Results from Two Phase I Trials (ALKA-372-001 and STARTRK-1). Cancer Discov 2017;7:400-9. [Crossref] [PubMed]
- Drilon A, Siena S, Dziadziuszko R, et al. Entrectinib in ROS1 fusion-positive non-small-cell lung cancer: integrated analysis of three phase 1-2 trials. Lancet Oncol 2020;21:261-70. [Crossref] [PubMed]
- Roys A, Chang X, Liu Y, et al. Resistance mechanisms and potent-targeted therapies of ROS1-positive lung cancer. Cancer Chemother Pharmacol 2019;84:679-88. [Crossref] [PubMed]
- Morris TA, Khoo C, Solomon BJ. Targeting ROS1 Rearrangements in Non-small Cell Lung Cancer: Crizotinib and Newer Generation Tyrosine Kinase Inhibitors. Drugs 2019;79:1277-86. [Crossref] [PubMed]
- Yun MR, Kim DH, Kim SY, et al. Repotrectinib Exhibits Potent Antitumor Activity in Treatment-Naïve and Solvent-Front–Mutant ROS1-Rearranged Non–Small Cell Lung Cancer. Clin Cancer Res 2020;26:3287-95. [Crossref] [PubMed]
- Davare MA, Vellore NA, Wagner JP, et al. Structural insight into selectivity and resistance profiles of ROS1 tyrosine kinase inhibitors. Proc Natl Acad Sci U S A 2015;112:E5381-90. [Crossref] [PubMed]
- Chong CR, Bahcall M, Capelletti M, et al. Identification of Existing Drugs That Effectively Target NTRK1 and ROS1 Rearrangements in Lung Cancer. Clin Cancer Res 2017;23:204-13. [Crossref] [PubMed]
- Dagogo-Jack I, Shaw AT. Expanding the Roster of ROS1 Inhibitors. J Clin Oncol 2017;35:2595-7. [Crossref] [PubMed]
- Drilon A, Ou SI, Cho BC, et al. Repotrectinib (TPX-0005) Is a Next-Generation ROS1/TRK/ALK Inhibitor That Potently Inhibits ROS1/TRK/ALK Solvent- Front Mutations. Cancer Discov 2018;8:1227-36. [Crossref] [PubMed]
- Cui J, Zhai D, Deng W, et al. TPX-0005, a Novel ALK/ROS1/TRK Inhibitor, Effectively Inhibited a Broad Spectrum of Mutations Including Solvent Front ALK G1202R, ROS1 G2032R and TRKA G595R Mutants. Eur J Cancer 2016;69:S32. [Crossref]
- Sehgal K, Piper-Vallillo AJ, Viray H, et al. Cases of ROS1-rearranged lung cancer: when to use crizotinib, entrectinib, lorlatinib, and beyond? Precis Cancer Med 2020;3:17. [Crossref] [PubMed]
- Killock D. LorlatinibinROS1-PositiveNSCLC. Nat Rev Clin Oncol 2020;17:7.
- Nicholas JR, Esmond DN, Janet LE, et al. Biomarker tissue journey among patients (pts) with untreated metastatic non-small cell lung cancer (mNSCLC) in the U.S. Oncology Network community practices. J Clin Oncol 2021;39:9004. [Crossref]
- Salas C, Martín-López J, Martínez-Pozo A, et al. Real-world biomarker testing rate and positivity rate in NSCLC in Spain: Prospective Central Lung Cancer Biomarker Testing Registry (LungPath) from the Spanish Society of Pathology (SEAP). J Clin Pathol 2022;75:193-200. [Crossref] [PubMed]
- Rossi G, Russo A, Tagliamento M, et al. Precision Medicine for NSCLC in the Era of Immunotherapy: New Biomarkers to Select the Most Suitable Treatment or the Most Suitable Patient. Cancers (Basel) 2020;12:1125. [Crossref] [PubMed]
- Rijavec E, Coco S, Genova C, et al. Liquid Biopsy in Non-Small Cell Lung Cancer: Highlights and Challenges. Cancers (Basel) 2019;12:17. [Crossref] [PubMed]
Cite this article as: Parisi F, Rossi G, Biello F, Tagliamento M, Barletta G, Zullo L, Cella E, Sacco G, Dellepiane C, Bennicelli E, Favero D, Alama A, Coco S, Marconi S, Zinoli L, Tanda ET, Pronzato P, Rijavec E, Genova C. Current state of the art on the diagnosis and the role of target therapy for treatment of ROS1-rearranged non-small cell lung cancer: a narrative review. Precis Cancer Med 2022;5:25.

