Clinical implications of liquid biopsies in urogenital tumors: a narrative review
Introduction
The current trends in cancer management are moving toward precision medicine and personalized therapy. Advances in this regard are being made on several fronts in oncology, from screening and early detection, to diagnosis, prognostication, treatment selection, response monitoring and detection of recurrence (1). Precision and personalized medicine aims to provide an individualized approach to treatment, tailored according to disease biology and predicted response (2). Liquid biopsy is a minimally invasive method that detects tumor biomarkers in different types of body fluids (blood, urine, sputum, saliva, cerebrospinal fluid, pleural effusion, etc.). Although its processing may be more technologically complex and it has a shorter half-life that traditional biopsy, liquid biopsy is associated with fewer complications than tissue biopsies and is easier to sample which allows for more frequent real-time monitoring of disease evolution at different time points (1). Because the test detects circulating biomarkers, it allows a better representation of inter and intra-tumor heterogeneity, which are contributors to treatment failure (3,4). Furthermore, liquid biopsies may be less costly and require a shorter processing time compared to tissue biopsies (4).
Urogenital malignancies are among the areas where the utility of liquid biopsy is being explored and evidence is emerging on its application in the diagnosis, prognosis and disease response monitoring in these types of tumors. In this review, we present an update on the available literature pertaining to the clinical utility of liquid biopsy in urogenital tumors, in particular bladder cancer (BC) and prostate cancer (PC), which are commonly encountered malignancies. We present the following article in accordance with the Narrative Review reporting checklist (available at https://pcm.amegroups.com/article/view/10.21037/pcm-22-1/rc).
Methods
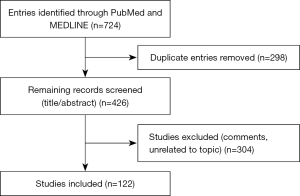
We searched the online databases PubMed and MEDLINE up to November 2, 2021, using the following Medical Subject Headings (MeSH) terms: “liquid biopsy”, “circulating tumor cells”, “exosomes”, “urogenital neoplasms”, “bladder neoplasms” and “prostate neoplasms”. There were no restrictions to language, and individual articles were screened to retrieve relevant studies. Articles discussing the clinical application of liquid biopsy in PC and BC detection, response to treatment, and prognosis were included. We excluded articles discussing preclinical testing of liquid biopsy technics. Tables 1,2 summarize the search strategy, and Figure 1 summarizes the literature search results.
Table 1
| Items | Specification |
|---|---|
| Date of search | November 2, 2021 |
| Databases and other sources searched | PubMed, MEDLINE, individual articles screening |
| Search terms used | “Liquid biopsy”, “circulating tumor cells”, “exosomes”, “urogenital neoplasms”, “bladder neoplasms” and “prostate neoplasms” |
| Timeframe | Up to November 2, 2021 |
| Inclusion and exclusion criteria | Review articles, observational, correlative and randomized studies were included. No language restriction |
| Selection process | Two of the authors independently conducted the literature search to select articles discussing the clinical application of liquid biopsy in PC and BC |
PC, prostate cancer; BC, bladder cancer.
Table 2
| Items | Specification |
|---|---|
| Date of search | November 2, 2021 |
| Databases and other sources searched | MEDLINE |
| Search terms used | (“Liquid Biopsy/” OR “Exosomes/” OR “Circulating Tumor DNA/” OR “Neoplastic Cells, Circulating/”) AND (“Urogenital Neoplasms/” OR “Prostatic Neoplasms/” OR “Urinary Bladder Neoplasms/”) |
| Timeframe | 1946 to November 2, 2021 |
| Inclusion and exclusion criteria | Review articles, observational, correlative and randomized studies were included. No language restriction |
| Selection process | Two of the authors independently conducted the literature search to select articles discussing the clinical application of liquid biopsy in PC and BC |
PC, prostate cancer; BC, bladder cancer.
Overview of liquid biopsy technics
Liquid biopsy encompasses several modalities, including circulating tumor cells (CTCs), cell-free circulating nucleic acids [circulating tumor DNA (ctDNA) or RNA], microvesicles, exosomes and tumor-educated platelets. Most of the studies that evaluated liquid biopsy in PCs and BCs employed CTC enumeration and phenotyping, cell free DNA (cfDNA) profiling and exosomal RNA.
CTCs are cells that detach from the primary tumor site or metastasis site and reach the blood (5). To survive in bloodstream, CTCs lose their epithelial characteristics and acquire a mesenchymal phenotype in a process termed epithelial-mesenchymal transition (EMT). EMT eases the metastatic spread of tumor cells and allows them to escape apoptosis, cell senescence and immune system (4,6).
Multiple technics for CTC detection and enumeration exist. CellSearch is the only FDA approved CTC detection method. Magnetic beads coated with anti-epithelial cell adhesion molecule (EpCAM) antibodies capture CTCs, which are further characterized based on morphology and staining [4',6-diamidino-2-phenylindole (DAPI)-positive nucleus, cytokeratin positive cytoplasm and CD45 negative] (7). The CTC count is expressed as the number of cells meeting these criteria per 7.5 mL of blood, which implies a relatively low sensitivity owing to the small amount of blood used (8). Only viable cells are counted in this technic and cellular fragments, anucleated cells and necrotic cells are excluded by the definition criteria for CTC using CellSearch (9). CellCollector overcomes the low sensitivity limitation by isolating CTCs in vivo in a larger blood volume, using a sterile wire with a functional tip coated with anti-EpCAM antibodies, inserted into the patients’ cubital vein. The EPithelial Immuno SPOT (EPISPOT) assay, can differentiate between viable and apoptotic CTCs, unlike the other two previously mentioned tests. EPISPOT detects proteins released or expressed by epithelial cancer cells, which are in turn captured on a membrane coated with antibodies against these proteins and detected by secondary labeled antibodies (10). It is important to note that studies should be interpreted in the light of the CTC enumeration method they utilized, for example EpCAM-dependent methods might fail to identify cells that have undergone EMT, which tend to be more aggressive (11).
Circulating cfDNA is released into the bloodstream from cells undergoing apoptosis or necrosis or actively from living cells through extracellular vesicles (4). Not all cfDNA originates from cancer cells and can be derived from blood cells or others (5). CtDNA is the portion of cfDNA that is derived from tumor cells. Highly sensitive technics such as droplet digital polymerase chain reaction (ddPCR) and next-generation sequencing (NGS) are required to identify ctDNA by specific genetic alterations within total cfDNA, which allows real time profiling of tumors (11). Figure 2 illustrates CTC and ctDNA technics in liquid biopsy.
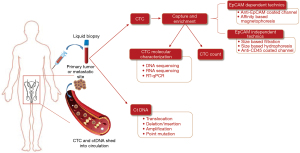
Exosomes are membrane-bound vesicles of 30–150 nm produced and secreted from cells into the extracellular space (12). They are implicated in several physiological and pathological processes (13). Exosomes play a significant role in organizing intercellular communication by transferring cargo between cells. Moreover, these nanoparticles are proven to be widely involved in oncogenesis and cancer progression (14). All exosomes are composed of specific proteins, lipids, messenger RNAs (mRNAs), and microRNAs (miRNAs) (15). Their abundance in body fluids turned them into a source of several potential cancer biomarkers (16). Thus, over the past years, exosomes have become a primary source for liquid biopsy, and their role in cancer care have been studied in multiple malignancies, including prostate and bladder neoplasms (16-18).
Liquid biopsy in PC
PC is the most common malignancy in males, with around 248,530 new cases yearly, and fifth leading cause of cancer death worldwide (5). The prognosis and treatment modalities depend on the stage of disease, from localized to metastatic, and serum testosterone that defines non-castrate or castrate state (8). The diagnosis is established by histopathologic examination of biopsy tissues, and prognosis relies on the biopsy Gleason score and prostate specific antigen (PSA) level. Genetic biomarker tests performed on tissue biopsies have been developed to stratify the risk of patients. For example, Oncotype DX detects the expression of 12 genes that play a role in tumorigenesis by reverse transcription PCR (RT-PCR) assay. The combination of results is used to calculate the Genomic Prostatic Score, which predicts unfavorable PC pathology (19). Another genomic test is Prolaris, which measures the expression of 46 genes that correlate with PC progression (20).
Current detection tools for PC have their disadvantages. Digital rectal exam can be limited by low compliance and dependency on the examiner’s expertise, and usually allows the detection of advanced stage disease. PSA is a protease secreted by prostate cells, and thus is not specific to cancer and can be elevated in other benign causes such as benign prostate hypertrophy (BPH), infection or infarction (5). For example, less than one third of patients with elevated PSA or abnormal digital rectal examination (DRE) turn out to have PC on transrectal ultrasound-guided biopsy (21). In another study, 40% to 50% of patients with high PSA had a biopsy-proven PC (5). Therefore, an alternative method with a better diagnostic capacity is needed. The clinical applications of liquid biopsy in metastatic castrate resistant PC (mCRPC) and in localized PC (LPC) and metastatic hormone sensitive PC (mHSPC) are summarized in Tables 3,4.
Table 3
| Study | Clinical application | Technics used | Population (N) | Objectives | Results |
|---|---|---|---|---|---|
| Sánchez et al. (22) | Diagnosis | Analysis of AR-CAG repeats in blood and PPM of GSTP1 and RASSF1A genes in prostate tissue, urinary sediment cells and plasma DNA using quantitative methylation-specific PCR | 186 patients scheduled for prostate biopsy | Combined analysis of AR-CAG repeat length and the PPM of CSTP1 and RAASF1A genes as a noninvasive assessment tool for PC | The combined analysis of blood biomarkers and urinary sediments able to differentiate PC from BPH |
| McKiernan et al. (23) | Diagnosis | ExoDx Prostate IntelliScore urine exosome assay | 1,563 participants with suspicious DRE/PSA level scheduled for biopsy | Discriminatory ability of urine exosome gene expression assay plus SOC vs. SOC alone | Urine exosome gene expression assay plus SOC associated with better discrimination between GS7 or greater and GS6 and benign disease |
| Danila et al. (24) | Prognosis | CTC enumeration in blood using CellSearch | 120 patients with mCRPC | Association of baseline CTC count with clinical characteristics and survival in patients with mCRPC | Strong correlation of baseline CTC count with survival with no threshold effect. No correlation between absence of CTCs and better outcome |
| de Bono et al. (25) | Prognosis | CTC enumeration in blood using CellSearch | 231 patients with mCRPC | Relation between post-treatment CTC count and OS | Unfavorable CTC count (≥5 CTC/7.5 mL) significantly associated with shorter OS and improvement of prognosis with conversion from unfavorable count to favorable |
| Scher et al. (26) | Prognosis | CTC enumeration in blood using CellSearch | 164 patients with mCRPC | Prognostic value of CTC count in patients with mCRPC receiving first line chemotherapy | Baseline LDH, baseline CTC and post-treatment fold change in CTC count predictive of survival |
| Huang et al. (27) | Prognosis | Plasma exosomal RNA sequencing with qRT-PCR | 100 patients with CRPC | Prognostic value of exosomal miRNAs in CRPC | Significant association of miR-1290 and miR-375 with poor OS |
| Del Re et al. (28) | Prognosis/treatment response | Plasma exosomal RNA extraction for analysis of AR-V7 by ddPCR | 36 patients with CRPC | Value of AR-V7 as predictive of resistance to hormonal therapy | Significantly worse PFS and shorter OS for AR-V7 positive patients |
| De Laere et al. (29) | Prognosis | CTC enumeration in blood using CellSearch, CTC-ARV targeted RNA-sequencing and sequencing of plasma cfDNA for AR and TP53 | 168 patients with mCRPC | Prognostic value of AR and TP53 profiling in patients with mCRPC starting a new line of ARSi | Outperformance of TP53 inactivation as indictor of negative prognosis for second-generation ARSi compared to any AR-derived biomarker |
| Mehra et al. (30) | Treatment response | Isolation of plasma cfDNA using QIAamp Circulating Nucleic Acid Kit | 571 patients with mCRPC | Evaluation of cfDNA concentration as prognostic marker after first- and second-line taxane chemotherapy | Baseline cfDNA concentration independent prognostic factor for rPFS and OS |
| Goodall et al. (31) | Prognosis/treatment response | Plasma cfDNA isolation using the QIASymphonyTM (Qiagen) and the circulating DNA kit (Qiagen) and quantification by Quant-iT High Sensitivity Picogreen Kit (Invitrogen) | 49 patients with CRPC | Prognostic value of changes in cfDNA concentration and predictive value of changes in cfDNA somatic mutation allele frequency in response to therapy | Decline of allele frequency of somatic mutations in responders to olaparib and a >50% decrease in in cfDNA after 8 weeks of therapy independently associated with longer OS |
| Torquato et al. (32) | Treatment response | CfDNA analysis using NGS for sequence variants and copy number gain in AR and 45 other cancer-related genes | 62 patients with mCRPC | AR gene alterations as markers of enzalutamide and abiraterone resistance | High ctDNA concentration associated with worse PSA response, PFS and OS. TP53 loss and PI3K pathway defects associated with worse OS |
| Okegawa et al. (33) | Treatment response | CTC enumeration in blood using CellSearch | 57 patients with bone mCRPC | Predictive value of CTC count in patients with bone mCRPC treated with docetaxel chemotherapy | Alkaline phosphatase and CTC count independent predictors of OS. CTC count ≥5 after 3 courses of chemotherapy associated with shorter OS |
| Vogelzang et al. (34) | Treatment response | CTC enumeration in blood using CellSearch | 208 patients with mCRPC | Prognostic value of CTC count in patients with mCRPC receiving docetaxel and lenalidomide | CTC count of ≥5 significantly associated with shorter OS. Shorter OS predicted by conversion of the CTC count from <5 to ≥5 after 3 cycles |
| Goldkorn et al. (35) | Treatment response | CTC enumeration in blood using CellSearch | 263 patients with mCRPC | Prognostic value of CTC count for OS in patients with mCRPC treated with first line docetaxel-based regimen | Significant association of CTC counts of <5 and ≥5 with OS (26 vs. 13 months, respectively). CTC count increase as a continuous variable associated with shorter OS |
| Heller et al. (36) | Treatment response/surrogate for survival | CTC enumeration in blood using CellSearch | 6,081 patients with mCRPC in five prospective trials | Early post-treatment decline in CTC count as indicator of prolonged survival for use in early-phase clinical trials | Highest discriminatory power for OS with CTC count decline from baseline nonzero to zero at week 13 and CTC conversion from unfavorable to favorable count |
| Lorente et al. (37) | Treatment response | CTC enumeration in blood using CellSearch | 511 patients with mCRPC and baseline CTC count <5/7.5 mL | Increase in CTC count as marker of disease progression in mCRPC patients with low baseline count treated with abiraterone or chemotherapy | Worse OS associated with CTC progression during the first 12 weeks of treatment |
| Scher et al. (38) | Surrogate for survival | CTC enumeration in blood using CellSearch | 711 patients with mCRPC | CTC count as a surrogate outcome measure in COU-AA-301 trial (abiraterone plus prednisone vs. prednisone alone) | Panel of LDH level and CTC count found to be a surrogate for survival at individual-patient level |
| Kienel et al. (39) | Prognosis/treatment response | Plasma cfDNA isolation using QIAamp Circulating Nucleic Acid kit | 59 patients with mCRPC | Prognostic value of cfDNA in patients with mCRPC who received first and second line chemotherapy | cfDNA strong independent predictor of OS and PSA response |
| Annala et al. (40) | Treatment response | CfDNA analysis with whole exome sequencing and deep targeted sequencing | 202 patients with mCRPC | Determination of genomic drivers of resistance to AR-directed therapy in mCRPC receiving enzalutamide or abiraterone | Strong correlation between defects in BRCA2 and ATM and poor outcomes. Somatic alterations in TP53 independently associated with rapid resistance |
| Conteduca et al. (41) | Treatment response | AR copy number and mutation detection in plasma DNA using ddPCR | 2 cohorts of patients with CRPC: primary cohort (treated with abiraterone or enzalutamide before or after docetaxel); PREMIERE cohort (chemo-naïve patients treated with enzalutamide) | Clinical qualification of AR gene status in plasma DNA pre and post chemotherapy | AR CN gain before starting enzalutamide or abiraterone correlated with decreased OS and PFS independent of prior chemotherapy status |
| Antonarakis et al. (42) | Treatment response | CTC detection using modified AdnaTest platform. AR-V7 detection in CTCs using quantitative PCR | Patients with mCRPC, 31 starting enzalutamide and 31 abiraterone | Relation between AR-V7 mRNA in CTC and resistance to enzalutamide and abiraterone | AR-V7 detection associated with lower PSA response rates, shorter PSA PFS, clinical or radiologic PFS and OS |
| Antonarakis et al. (43) | Treatment response | CTC detection using modified AdnaTest platform. AR-V7 detection in CTCs using quantitative PCR | 202 patients with CRPC | Prognostic value of AR-V7 mRNA detection in CTCs in patients treated with abiraterone or enzalutamide | Best outcomes seen in patients with negative CTC count, followed by those with CTC positive/AR-V7 negative, followed by those with CTC positive/AR-V7 positive |
| Scher et al. (44) | Treatment response | Epic Sciences CTC nuclear-specific AR-V7 protein assay | 142 patients with mCRPC | Nuclear localization of AR-V7 protein in CTCs as determinant of OS in patients treated with taxanes vs. ARSi | Higher OS in high-risk patients positive for AR-V7 and treated with taxanes compared to those who received ARSi. Improved OS in patients negative for AR-V7 treated with ARSi compared to taxanes (19.8 vs. 12.8 months, P=0.05) |
| Darshan et al. (45) | Treatment response | CTC detection in blood using CellSearch enrichement or Ficoll separation. AR detection with immunofluorescence | 14 patients with CRPC | Mechanisms of taxane inhibition of AR, and AR localization in CTCs as predictor of response to chemotherapy | 30% increase in odds of responding to taxane with complete AR localization in the cytoplasm vs. no AR in cytoplasm |
| Salvi et al. (46) | Treatment response | Serum DNA isolation and CNVs analysis using Taqman copy number assays | 53 patients with CRPC | Evaluation of CNVs of CYP17A1 and AR genes in cfDNA before starting abiraterone | Shorter PFS associated with AR and CYP17A1 CNVs and PSA decline ≥50%. Performance status, PSA decline ≥50%, AR CNV and DNA concentration correlated with OS |
| Armstrong et al. (47) | Prognostic/treatment response | Modified-AdnaTest CTC AR-V7 mRNA assay and Epic Sciences CTC nuclear-specific AR-V7 protein assay | 118 patients with mCRPC | Validation of poor prognostic value of AR-V7 detection in patients treated with abiraterone or enzalutamide | AR-V7 detection by both methods correlated with shorter PFS and OS, independently of CTC count and clinical prognostic factors |
mCRPC, metastatic castrate resistant prostate cancer; AR, androgen receptor; PPM, percentage of promoter methylation; GSTP1, glutathione S-transferase Pi 1; RASSF1A, RAS association domain family 1A; PCR, polymerase chain reaction; CTC, circulating tumor cell; qRT-PCR, quantitative reverse transcription polymerase chain reaction; AR-V7, androgen receptor splice variant 7; ddPCR, droplet digital polymerase chain reaction; cfDNA, cell free DNA; TP53, tumor protein 53; NGS, next-generation sequencing; DRE, digital rectal exam; PSA, prostate-specific antigen; PC, prostate cancer; SOC, standard of care; OS, overall survival; miRNA, microRNA; ARSi, androgen receptor signaling inhibitor; mRNA, messenger RNA; CNV, copy number variation; BPH, benign prostatic hyperplasia; GS, Gleason score; LDH, lactate dehydrogenase; PFS, progression-free survival; rPFS, radiologic progression-free survival; ctDNA, circulating tumor DNA; PI3K, phosphoinositide 3-kinase; BRCA2, breast cancer gene 2; ATM, ataxia telangiectasia mutated; CN, copy number; CYP17A1, cytochrome P450 17A1.
Table 4
| Study | Clinical application | Technics used | Population (N) | Objectives | Results |
|---|---|---|---|---|---|
| Patients with HSPC | |||||
| Goodman et al. (48) | Prognostic/treatment selection | CTC enumeration in blood using CellSearch | 33 patients with HSPC receiving ADT | Clinical utility of CTC enumeration in HSPC | Baseline CTC count independent predictive marker for progression to CRPC |
| Josefsson et al. (49) | Prognosis | CTC detection with AdnaGen Prostate Cancer Detect/Select kit. AR-V7 mRNA detection with qRT-PCR | 36 patients with mHSPC | Prognostic role of genes associated with development of CRPC | AR-V7 detection independent prognostic marker for cancer specific survival |
| Yang et al. (50) | Prognosis/treatment response | CTC enumeration using Can Patrol technic (detects markers of EMT) | 108 patients with mHSPC | Determination of markers predictive of response to ADT in patients with newly diagnosed mHSPC | Shorter time to CRPC in patients with mesenchymal phenotype CTCs compared to those with non-mesenchymal phenotype CTCs or negative CTCs |
| Patients with LPC | |||||
| Thalgott et al. (51) | Prognosis | CTC detection in blood using CellSearch | 15 patients with LAPC and 15 healthy controls | Prognostic value of CTC in detecting BCR in patients with LAPC undergoing neoadjuvant chemo-hormonal therapy and radical prostatectomy | Stochastic CTC detection and no correlation with clinic-pathological characteristics. It could predict BCR (not statistically significant) |
| Meyer et al. (52) | Prognosis | CTC detection in blood using CellSearch | 152 patients with LPC | Prognostic value of CTC detection in LPC before radical prostatectomy | Low CTC detection rate (11%) and no significant correlation with BCR, PSA or disease characteristics |
| Loh et al. (53) | Prognosis | CTC detection in blood using CellSearch | 36 patients with high-risk non-metastatic PC | Determination of the rate of CTC detection in patients with high risk LPC | No correlation between CTC detection and known prognostic factors (PSA, Gleason score, T-stage and age) |
| Davis et al. (54) | Prognosis | CTC detection in blood using CellSearch | 97 patients with LPC and 25 controls with high PSA but no cancer detected | Evaluation of CTC detection correlation with tumor volume, pathological stage and Gleason score in patients with LPC | Low CTC count and comparable to patients with no cancer diagnosis. No correlation between CTC count and other prognostic factors |
| Aragon-Ching et al. (55) | Prognosis | CTC detection in blood using CellSearch | 36 patients post primary treatment with prostatectomy or radiation | CTC measurability in patients with BCR and its predictive ability of PSA increase and PSA doubling times | Infrequent presence of CTCs in patients presenting with BCR, but increased suspicion for metastasis with CTC detection |
| Todenhöfer et al. (56) | Prognosis | Antigen-independent enrichment of CTC using microfluidic platform | 50 patients with LPC | Evaluation of CTC detection in patients with LPC by microfluid platform | CTC detection in 50% of patients, but no correlation with PSA, cancer stage, lymph node stage or Gleason score |
| Friedlander et al. (57) | Prognosis | CTC enumeration using Epic Sciences system | 37 patients with high risk LPC | Prognostic role of CTC in high risk LPC | Shorter recurrence time in patients with detectable CTCs (not statistically significant). Positive CTC count in all patients with BCR |
LPC, localized prostate cancer; mHSPC, metastatic hormone sensitive prostate cancer; CTC, circulating tumor cell; CRPC, castration-resistant prostate cancer; AR-V7, androgen receptor splice variant 7; mRNA, messenger RNA; qRT-PCR, quantitative reverse transcription polymerase chain reaction; EMT, epithelial-mesenchymal transition; ADT, androgen deprivation therapy; LAPC, locally advanced prostate cancer; PC, prostate cancer; PSA, prostate specific antigen; BCR, biochemical recurrence.
Role of liquid biopsy as PC detection tool
CfDNA was found to be elevated in patients with PC compared to those with PBH or healthy controls in different studies, with a sensitivity of 80% and a specificity of 82%, making it a diagnostic marker (1).
The methylation pattern of several genes in cfDNA found in urine or blood samples can detect early PC (5). Although some epigenetic alterations detected in circulating cfDNA such as methylated glutathione S-transferase pi 1 (GSTP1me) have high specificity for PC, multigene panels have been shown to improve the sensitivity of these tests. Ellinger et al. demonstrated that using a gene panel increased the sensitivity of diagnosing PC to 47%, compared to 42% by testing GSPT1me alone (58). In another study, a panel including GSTP1me and methylated RAS association domain family 1A (RASSF1Ame) had a 71% positive predictive value (PPV) and 59.6% negative predictive value (NPV) for cancer detection in a population of biopsy proven PC patients. These values increased when serum PSA was also taken into account, to reach a PPV of 81% and NPV of 66% (22).
Urinary biomarkers have also been shown to be better diagnostic tools than PSA with the ability to predict high-grade cancer, therefore reducing the number of unnecessary biopsies. There are 4 commercially available urinary biomarker tests: Progensa PC gene 3 (PCA3), SelectMDx, Michigan Prostate Score (MiPS) and ExoDx Prostate (IntelliScore). Longitudinal studies are needed to establish their clinical utility in guiding diagnostic and therapeutic decisions. Furthermore, their role in patients with advanced cancer is limited, as these patients often lack the primary cancer (59,60).
Prognostic value of liquid biopsy in metastatic PC
The detection of CTC reflects disease activity and spread, and is therefore associated with a worse prognosis in metastatic PC (61). Unlike PSA, CTC detection is not dependent on androgen receptor (AR)-mediated signaling pathways, making it a reliable marker of disease burden (5,8). Promising evidence exists supporting the use of CTC as a marker in mCRPC. When using the CellSearch assay, the detection of 5 or more CTCs per 7.5 mL is associated with an unfavorable prognosis, and this is the cut-off specified by the US FDA (5). In a study conducted by Danila et al. that detected CTCs using CellSearch in 120 patients with progressive clinical castrate metastatic disease, a decrease in survival was observed as CTC count increases. In fact, CTC count analyzed as a continuous variable was an independent prognostic factor for survival with no threshold effect, which contests the use of a discrete cutoff. In addition, CTC count moderately correlated with other determinants of disease severity like PSA and bone scan index. However, survival varied for patients with no detected CTCs, which implies that the absence of CTCs does not necessarily correlates with a better outcome (24). IMMC-38 was the most important trial to demonstrate the prognostic capacity of CTC in mCRPC. In this prospective, open-label study, CTC count was analyzed in patients with progressive CRPC before starting a new line of chemotherapy and monthly afterward. Patients with unfavorable baseline CTC count (≥5 CTC/7.5 mL) had lower overall survival (OS) compared to those with a favorable count. In addition, the highest OS was seen in patients who maintained a favorable CTC count throughout all points of evaluation (26 months), followed by patients who converted from unfavorable to favorable count (21.3 months), and the shortest OS was observed in patients whose count remained unfavorable (6.8 months). The difference in OS between these groups was statistically significant. This study also showed that patients who converted from favorable to unfavorable count after treatment had a poor prognosis (OS: 9.3 months), but the number of participants in this group was small (25). Reanalysis of the data from this study showed that baseline lactate dehydrogenase (LDH) and CTC evaluated as a continuous variable were independent prognostic factors of OS, unlike changes in PSA titers (26).
Another important consideration is that prior to first line therapy, only 25% of patients present with CTC number ≥5 cells per 7.5 mL. Therefore, more research is needed to identify a lower threshold, more suitable for establishing prognosis (5).
DNA and RNA profiling also has a prognostic role in metastatic PC. Huang et al. sequenced exosomal RNA in order to identify prognostic markers in patients with CRPC. MiR-1290 and miR-375 were associated with poor OS and are promising prognostic markers in CRPC (27). A urine exosome gene expression assay including V-ets erythroblastosis virus E26 oncogene homologs (ERG), PCA3 and SAM pointed domain-containing Ets transcription factor (SPDEF), was able to discriminate high-grade (Gleason score ≥7) from low-grade cancer (Gleason score 6) and benign disease in patients with elevated PSA. This assay has the potential to identify patients who require biopsy and thus decrease the number of unnecessary interventions (23).
In addition, mutations in breast cancer gene 2 (BRCA2), tumor protein p53 (TP53), ataxia telangiectasia mutated (ATM) and phosphatidylinositol-3-kinase (PI3K) identified in ctDNA were associated with a negative prognosis in patients with CRPC, independent of ctDNA concentration and other clinical prognostic factors (60,62).
The AR is a main contributor to PC development, progression and resistance to castration. Alterations of AR including AR amplification, point mutation and splice variants are hypothesized to cause CRPC. AR variants have lost the C-terminal domain where testosterone naturally binds, which results in a ligand-independent constitutional activation of the receptor. AR isoform encoded by AR splice variant 7 (AR-V7) is the most commonly and clinically significant variant associated with CRPC (42). AR-V7 can be detected in exosomal RNA, which is simpler and less expensive than CTC processing. Using this method, Del Re et al. found that AR-V7 positive patients had worse progression-free survival (PFS) and shorter OS (28).
Role of liquid biopsy in treatment choice and response evaluation
PSA is used as a biomarker to monitor response to treatment. However, because its genetic encoding is androgen-dependent, PSA levels may decrease in response to lowering of serum androgen level or direct effect of some therapeutic agents on the PSA gene, without corresponding to cytoreduction (5,8,60). Therefore more accurate biomarkers are needed to reflect treatment response and allow better tumor profiling to guide the choice of treatment (11).
Metastatic PC that progresses with androgen deprivation therapy (ADT) is termed castration resistant. Multiple alternative therapeutic options exist in this case, such as next generation AR targeting agents (e.g., abiraterone and enzalutamide), taxane chemotherapy (docetaxel and cabazitaxel) and other treatments like sipuleucel-T and radium-223. However, one quarter of patients develop de-novo resistance to AR targeting agents, and cross-resistance is common among this class. Therefore, there is an unmet need to predict treatment resistance in mCRPC (60). Baseline CTC count was found to be an independent predictor of progression to castrate resistance in 33 men with mHSPC, and therefore can help identify patients who would benefit from early chemotherapy (48).
Several trials have examined the prognostic role of CTC enumeration in predicting survival in mCRPC patients on chemotherapy, and the results agree that a baseline CTC count of ≥5 and a post-treatment CTC count of ≥5 are significantly associated with shorter OS (33-35).
In a pooled analysis including 6,081 patients with mCRPC from five prospective trials (COU-AA-301, AFFIRM, ELM-PC-5, ELM-PC-4, and COMET-1), Heller et al. studied the CTC count and PSA change at treatment week 13. They concluded that “CTC0” (CTC nonzero at baseline and zero at 13 weeks) and “CTC conversion” (≥5 CTCs at baseline, ≤4 at 13 weeks) endpoints had the highest discriminatory power for OS, and would make meaningful endpoints in clinical trials (36).
Lorente et al. performed a post hoc analysis of patients with advanced PC and baseline CTC <5 treated in the IMMC-38 trial (chemotherapy) and COU-AA-301 trial (abiraterone or placebo plus prednisone). They found that a rise in CTC count during the first 12 weeks of treatment was independently associated with worse OS (37). A secondary endpoint of the COU-AA-301 trial was to evaluate the potential of CTC count alone or with a panel with other biomarkers as a surrogate endpoint for survival. The results were that CTC count in combination with LDH satisfied the prentice criteria as a surrogate for survival at the individual patient level (38).
However, the use of CTC count as an assessment of treatment response in clinical trials does not automatically translate into clinical practice on individual patients. First, the rates of detecting CTC count ≥5 in mCRPC differ according to the line of treatment, and vary from 30% in first line setting to 50% in third line. This is an important consideration to take when screening patients to establish the efficacy of a certain treatment. Second, we lack prospective studies that demonstrate whether changing regimens based on unfavorable CTC count would prove to be clinically useful. Does failure to convert from unfavorable to favorable count necessarily dictate a change in therapy? As an example, if the CTC count drops from 100 to 10, it would still be considered in the unfavorable range, while a decrease from 6 to 4 is seen as a favorable conversion and keeping same therapy is warranted. This example illustrate that making therapeutic decisions cannot purely rely on CTC enumeration, the interpretation of which should be made with caution and in the context of a global clinical picture. In addition, there is insufficient data on the interpretation of fluctuations in CTC count throughout treatment, and it is not clear whether a single increase in CTC count is sufficient to predict poor prognosis or serial assessments are needed (8).
CfDNA quantification was evaluated across different studies to establish its correlation with disease burden and response to treatment. Kienel et al. showed that a concentration of cfDNA above a threshold of 55.03 ng/mL was significantly associated with poor PSA response to chemotherapy and that cfDNA was an independent predictor of OS [hazard ratio (HR): 0.34; 95% confidence interval (CI): 0.12–0.91; P=0.032] in multivariate analysis (39). Another study by Kwee et al. demonstrated that the concentration of cfDNA significantly increased across three time points in the study: before chemotherapy, after the first cycle and after the third cycle. Additionally, cfDNA correlated significantly to mean maximum standardized uptake value (SUVmax) on positron emission tomography/computed tomography (PET/CT) at all three time points but not with PSA level changes. Furthermore, changes in promoter methylation status of two cancer-related genes [GSTP1 and retinoic acid receptor B2 (RARB2)] in cfDNA were observed with the metabolic changes captured by PET/CT. However, the translational significance of these findings and their potential value as response biomarkers remains to be further investigated (63).
cfDNA allows the detection of aberrations of the AR gene, such as mutations and amplifications or splice variants, which can lead to resistance to ADT, and therefore aid in choosing more appropriate therapeutic options for those patients (5). Several studies have identified point mutations in the AR gene that are associated with resistance to abiraterone (such as L702H and T878A) and ezalutamide [c.2629T>C(F877L)]. However, Annala et al. demonstrated that AR gain in ctDNA is only associated with poor response to treatment if present in high copy number (≥8 copies), although this finding was not statistically significant in multivariate analysis (40,60).
CTC phenotyping also can identify molecular profiles of AR. AR-V7 is the first management predictive biomarker in PC; it is found in up to 75% of men with mCRPC and its expression increases after treatment with abiraterone or enzalutamide (6). Antonarakis et al. demonstrated that the detection of AR-V7 mRNA by RT-PCR in the CTCs of patients with mCRPC is associated with resistance to abiraterone or enzalutamide. Patients who were AR-V7 positive had lower PSA response rates, shorter PSA PFS, clinical or radiological PFS and OS compared to those who were negative (42). A later study by this group showed when combining CTC count and AR-V7 detection using a CTC-based mRNA assay, patients with mCRPC receiving novel hormonal therapy as first or second line can be stratified into three prognostic categories. CTC negative patients had the best outcomes, followed by CTC positive/AR-V7 negative patients, and lastly CTC positive/AR-V7 positive patients (43). Using the Epic Sciences CTC nuclear-specific AR-V7 protein assay, Scher et al. showed that in patients with mCRPC, the nuclear localization of AR-V7 protein in CTCs was related to lower OS in those treated with novel antiandrogens compared those treated with taxanes (7.3 vs. 14.3 months respectively, P=0.25) (44). These results are in line with a previous study, which showed that mCRPC patients had a 30% increased odd of responding to taxane if AR is localized completely in the cytoplasm compared to those with no AR in the cytoplasm (P value <0.001) (45).
Patients with CRPC were evaluated for AR and cytochrome P450 17A1 (CYP17A1) genes copy number variations (CNVs) in cfDNA before the start of abiraterone therapy. CYP17A1 is a key enzyme for sex steroid synthesis and a target of abiraterone. AR and CYP17A1 gene gain was associated with lower PFS and OS (46).
In the PROPHECY trial, a multicenter prospective study in men with high-risk mCRPC starting abiraterone or enzalutamide, AR-V7 detection in CTCs (using Johns Hopkins University modified-AdnaTest CTC AR-V7 mRNA assay and Epic Sciences CTC nuclear-specific AR-V7 protein assay) was found to be an independent predictive biomarker of short PFS and OS in this patient population (47).
Therefore the detection of AR-V7 in CTCs can be used as a biomarker to identify resistance to androgen pathway-targeted therapies, and the National Comprehensive Cancer Network (NCCN) guidelines recommend that the “use of AR-V7 tests can be considered to help guide selection of therapy in the post-abiraterone/enzalutamide metastatic CRPC setting” (64). However, there are certain limitations to consider, such as the heterogeneity of AR-V7 status among individual CTCs, the change in AR-V7 status that can occur during treatment, and the presence of alternative pathways of antiandrogen therapy that can contribute to clinical resistance (8).
Germline and somatic homologous recombination deficiency (HRD) is a target of several anticancer treatments, including chemotherapy and inhibitors of DNA repair enzyme [poly(ADP ribose) polymerase (PARP) inhibitors]. HRD biomarkers based on liquid biopsy have been developed. Targeted and whole-exome sequencing of cfDNA from patients in the TOPARP-A trial, a phase II trial of the PARP inhibitor olaparib in mCRPC, showed that allele frequency of somatic mutations declined in responders and that a decrease of ≥50% cfDNA concentration after 8 weeks of olaparib therapy was independently associated with longer OS (31). Mehra et al. demonstrated that cfDNA concentration is an independent prognostic factor for radiologic PFS and OS in patients with mCRPC receiving taxane chemotherapy in first and second line settings, and may serve as a biomarker of response to taxanes (30). In May 2020, olaparib received FDA approval for treatment of mCRPC in patients with germline or somatic homologous recombination repair (HRR) gene mutation who did not respond to enzalutamide or abiraterone. Approval was also granted to two blood-based tests FoundationOne CDx and BRACAnalysis CDx for detection of HRR gene alterations (65).
Role of liquid biopsy in early stage LPC
Fewer studies exist on the role of liquid biopsy in LPC and mHSPC. Thalgott et al. measured CTCs using CellSearch in plasma of patients with locally advanced PC before and after neoadjuvant chemotherapy and following subsequent radical prostatectomy. They found that CTC detection in these patients was random, and although it could predict biochemical recurrence in some cases, this finding was not statistically significant. In addition, the presence of CTCs did not correlate with like other clinicopathologic characteristics like initial Gleason score, serum PSA and TNM stage (51). Similar results were seen across other studies with different treatment modalities (52-54). The role of CTC in detecting biochemical recurrence and its correlation with PSA increase and PSA doubling time was also studied. This is particularly important because the assessment of biochemical recurrence in LPC by PSA can be variable, and these patients can benefit from further local salvage therapy and be offered a change for cure, while those who developed occult or overt metastasis can receive early systemic therapy. The incidence of positive CTCs was low (8.3%), however its presence raises the suspicion for metastasis (55).
The low yield of CTC detection in LPC can be attributed to the enumeration method used. The studies mentioned above used CellSearch, an EpCAM-dependent technic, which can underestimate the number of CTCs that underwent EMT and downregulated their epithelial markers (EpCAM and cytokeratins). Additionally, around 11% of prostate adenocarcinomas lack or have weak EpCAM expression (51,53). This idea is illustrated in the study by Kuske et al. that compared CTCs in patients with high-risk LPC before and after radical prostatectomy using 3 commercially available systems: 37% of the cohort had positive CTCs detected with CellSearch, 54.9% with CellCollector and 58.7% with EPISPOT. In addition, the CTC count measured by EPISPOT was significantly associated with serum PSA and the clinical cancer stage (66). These findings were corroborated by another study by Cieślikowski et al. (67). Other investigators have used label-independent platforms. One study used a microfluid platform to isolate CTCs based on cell size and deformability, in patients with LPC before radical prostatectomy. They were able to detect CTCs in 50% of the patients and showed no significant correlation between CTC count and clinicopathological parameters (56). Another study detected CTCs in 81.1% of patients with LPC after radical prostatectomy using label-independent Epic Sciences systems. Patients with higher CTC count tended to have shorter recurrence time, and although this was not statistically significant, the results are promising for the implementation of Epic Sciences platform for CTC detection as a prognostic marker in LPC (57).
Role of liquid biopsy in mHSPC
The data available on the prognostic value of liquid biopsies in mHSPC is scarce. In one study, ctDNA was detectable in patients with de novo mCSPC, however the levels decline after ADT), which renders the evaluation of clinically significant somatic mutations difficult. Therefore, if ctDNA were to guide therapeutic decisions in this case, the timing of blood collection is an important consideration (68). With respect to CTCs, a study by Josefsson et al. detected AR-V7 expression in CTCs of patients with mHSPC before the starting ADT. AR-V7 positivity was an independent negative prognostic marker for cancer specific survival in a multivariate analysis (49). Yang et al. showed that HSPC patients with mesenchymal phenotype CTCs had significantly higher progression rate to CRPC and shorter time to progression compared to patients with non-mesenchymal phenotype CTCs or absent CTCs (50).
Liquid biopsy in BC
BC is a common cancer worldwide with an estimated 573,278 new cases and 212,536 deaths in 2020 (69). It constitutes the 10th most common cancer globally and ranks 12th among the causes of cancer death. BC can present as muscle-invasive in 10–25% of patients, whereas the majority have non-muscle invasive disease (70). The treatment differs between the two subtypes. According to the NCCN guidelines, muscle-invasive BC (MIBC) patients are initially treated with cisplatin based neoadjuvant chemotherapy followed by radical cystectomy (71). On the other hand, the primary treatment of the supposed non-muscle-invasive BC (NMIBC) is a complete transurethral resection of the bladder tumor (TURBT) (71). However, recurrence is common among the NMIBC population, and approximately 10–30% of this group will experience progression toward a more aggressive disease (72).
The main challenge in all cancer cases lies in early detection and diagnosis before disease progression. Over the past decades, the diagnosis of BC consisted of cystoscopy and cytology. However, given the poor sensitivity of cytology and the high invasiveness of cystoscopy, it was crucial to channel the efforts toward some new diagnostic modalities that would provide new clinical understanding of the disease (73). Researchers and clinicians describe different levels of heterogeneity in BC. The tumor is characterized by high rates of somatic mutations, and genomic instability (74). Heide et al. highlighted the genomic heterogeneity of the tumor by implementing multiregional whole-exome sequencing from ten whole cystectomy specimens of BC (75). The sequencing showed key molecular alterations unevenly distributed over different areas of the primary tumor (75). This experiment supports the idea that a single tissue biopsy cannot be representative of all mutations of the entire tumor. Similarly, this intra-tumoral heterogeneity raises the issue of probable inter-tumoral differences between the primary cancerous site and metastases. Moreover, MIBC is divided into different subtypes based on transcriptome heterogeneity, i.e., gene expression profiling (76,77). These different subtypes can be recognized by the expression of specific biomarkers that could influence their response to certain targeted therapies (77).
For this purpose, in the past 20 years, precision oncology focused on the use liquid biopsy as a new method for analyzing tumor biomarkers in different body fluids, including blood, saliva, and urine (78,79). Recently, the concept of liquid biopsy has been gradually emerging in BC. By overcoming the temporal and spatial limitation of tissue biopsy, this new technic may play an essential role in the revolutionary movement of diagnosis, surveillance, prognosis and management of the disease (21,80-82). Table 5 summarizes the clinical applications of liquid biopsy in BC.
Table 5
| Study | Cancer type | Clinical application | Technics used | Population (N) | Objectives | Results |
|---|---|---|---|---|---|---|
| Grivas et al. (83) | Metastatic UC | Prognosis/therapeutic | Plasma cfDNA analysis using a 73-gene sequencing panel (Guardant360) | 124 patients with locally advanced or metastatic UC | Correlation between specific molecular alterations and role in prognosis and treatment of patients with aUC | Alterations in BRCA1 and RAF1 associated with shorter OS and FFS. Potential role of RAF kinase inhibitors proposed for treatment of aUC |
| Sundahl et al. (84) | Treatment response | Plasma ctDNA analysis, sequencing, quantification and calculation of ctDNA fraction | 18 patients with mUC divided into 2 arms | Assessment of pembrolizumab safety when combined with SBRT and evaluation of ctDNA fraction changes in concordance with treatment | Rapid decline of ctDNA fraction in responding patients, with stable or increased fraction in non-responders | |
| Vandekerkhove et al. (85) | Prognosis | Profiling plasma cfDNA through targeted sequencing, and calculation of ctDNA fraction | 104 patients with mUC and 39 patients initially diagnosed with local or locally advanced MIBC | Prognostic value of plasma ctDNA profiling vs. tumor tissue sequencing in patients with mUC | CtDNA abundance predictive of aggressiveness in MIBC patients, and highly prognostic for outcome in mUC | |
| Gallagher et al. (86) | Prognostic | CTC enumeration in blood using CellSearch | 33 patients with mUC | Correlation between CTC number and clinical parameters (metastatic sites, extent of disease burder) | CTCs detected in 44% of patients, and higher count in those with 2 or more metastatic sites. CTC count as potential early marker of response to chemotherapy | |
| Chen et al. (87) | UC with LN metastasis | Prognosis | Examination of exosomal LNMAT2-binding proteins using an RNA pull-down assay | Cohort 1: 266 patients with BC. Cohort 2: 206 BC patients and 120 healthy individuals | BC cell-secreted exosome-mediated lymphangiogenesis enhanced LN metastasis in BC patients independently of VEGF-C | LNMAT2 overexpression in BC tissues and enrichment in urinary exosomes correlated with LN metastasis |
| Okegawa et al. (88) | Metastatic UC and non-metastatic UC | Prognosis | CTC enumeration in blood using CellSearch | 20 patients with metastatic UC and 16 patients with non-metastatic UC | Usefulness of CellSearch system in detecting UC cell lines, and the correlation between CTC counts and UC clinical parameters | Higher CTC count in patients with more than one-site metastasis → correlation between CTC and disease burden |
| Rink et al. (89) | Localized UC and metastatic UC | Prognosis | CTC enumeration in blood using CellSearch | 50 patients with localized BC and 5 patients with metastatic disease | Significance of CTC in patients with non-metastatic advanced BC | Significantly worse OS, PFS and cancer-free survival in CTC-positive patients compared to those with negative CTC count |
| Abrahamsson et al. (90) | aUC | Prognosis | CTC enumeration in blood using CellSearch and further assesment for expression of EpCAM | 88 patients with histopathologically confirmed UC | Role of CTC as possible biomarker for disseminated disease in patients with advanced BC | Significant association of CTC presence with metastatic disease on FDG-PET-CT and increased risk of progression among patients treated with RC with or without perioperative chemotherapy |
| Christensen et al. (91) | NMIBC | Prognosis | FGFR3 and PIK3CA mutation detection in ctDNA from urine, plasma and tissue samples using ddPCR assays. CfDNA isolation from urine and plasma using the QIASymphony Circulating NA Kit | 363 patients with NMIBC and 468 patients with BC undergoing RC | Value of hotspot FGFR3 or PIK3CA mutations in BC surveillance | CtDNA levels in urine supernatants correlated with disease progression (NMIBC cohort). High level of urinary ctDNA associated with disease recurrence |
| Zhang et al. (92) | Prognosis | Alterations in tumor tissue DNA and plasma ctDNA detected using targeted sequencing based on an 861-gene panel | 82 NMIBC patients who underwent TURB followed by intravesical BCG | Evaluation of NGS use in screening plasma ctDNA for somatic alterations in patients with NMIBC | CtDNA detection rate and concordance between plasma and tissue somatic DNA variations associated with tumor size. High mTBI independent predictor of recurrence | |
| Guzzo et al. (93) | Prognosis | CTC enumeration in blood using CellSearch | 43 patients with UC who underwent RC | Role of CTC in predicting pathologic stage in patients with UC undergoing RC | Low numbers of CTC detected in a small percentage of patient → CTC CellSearch not likely useful in predicting extravesical disease | |
| Soave et al. (94) | Prognosis | CTC enumeration in blood using CellSearch | 226 patients with UC treated with RC without neoadjuvant chemotherapy | Evaluation of outcomes in patients with UC treated with RC according to CTC presence ± adjuvant chemotherapy | Presence of CTCs associated with disease recurrence, significant cancer-specific and overall mortality in patients not receiving adjuvant chemotherapy | |
| Kim et al. (95) | Prognosis | Urinary levels of cell free miR-214 quantification using real-time PCR | 138 patients with primary NMIBC and 144 healthy individuals | Evaluation of miR-214 as potential prognostic biomarker for NMIBC | Higher levels of miR-214 in patients with NMIBC vs. controls. MiR-214 level independent predictor of NMIBC recurrence | |
| Jiang et al. (96) | Diagnosis/prognosis | Genome wide analysis of serum miRNA using Miseq sequencing and SYBR PrimeScript miRNA RT-PCR Kit (Takara Bio) | 250 patients with BC and 240 controls | Serum miRNA expression in patients with BC as biomarker for early diagnosis and prognosis | Development of a six-miRNA diagnostic panel with sensitivities significantly higher than those of urine cytology. MiR-152 independent predictor of tumor recurrence | |
| Du et al. (97) | Diagnosis and prognosis | Genome wide analysis of serum miRNA using Miseq sequencing and SYBR PrimeScript miRNA RT-PCR Kit (Takara Bio) | 276 BC patients and 276 control individuals | Development of a urinary miRNAs panel for diagnosis and disease recurrence prediction in patients with BC | Development of seven-miRNA panel with high diagnostic accuracy and significantly higher AUCs compared to those of urine cytology. MiR-22-3p and miR-200a-3p independently associated with RFS of NMIBC | |
| Sapre et al. (98) | Prognosis | Urine total RNA extraction using mirVana miRNA Isolation Kit. Reverse transcription performed using the Taqman microRNA Reverse Transcription Kit | Screening phase of 81 patients (21 controls, 30 with active cancer and 30 non-recurrers). Validation of miRNA signature in 50 patients (25 with cancer and 25 without) | Evaluation of urine miRNA panel profiling as biomarker for recurrence of BC during surveillance phase | A six-miRNA panel best to predict disease recurrence, and was validated in independent cohort (AUC =0.74) | |
| Nicolazzo et al. (99) | Treatment selection | CTC enumeration in blood using CellSearch and ScreenCell. Testing CTCs for PD-L1 expression | 20 patients with NMIBC | Association between CTC expression of PD-L1 and response to BCG in NMIBC patients | PD-L1 positive CTCs in 92% of the BCG-unresponsive patients, and might guide targeted therapy after BCG failure | |
| Jiang et al. (100) | NMIBC and MIBC | Prognosis | Genome wide analysis of serum miRNA using Miseq sequencing and SYBR PrimeScript miRNA RT-PCR Kit (Takara Bio) | 207 patients with MIBC, 285 patients with NMIBC and 193 controls | Presence of stage-specific serum miRNAs and their potential use in predicting MIBC | Development of a 4 miRNAs MIBC detection panel with a significantly higher sensitivity and specificity than those of grade and urine cytology. MiR-486-3p and miR-103a-3p independently associated with OS |
| Rink et al. (101) | Non-metastatic UC | Prognosis | CTC enumeration in blood using CellSearch | 120 clinically non-metastatic UC patients | Predictive ability of CTC in advanced disease stages, extravesical nodal status and outcome after RC | CTC associated with inferior disease outcomes |
| Rink et al. (102) | Prognosis | CTC enumeration in blood using CellSearch. Determination of CTC HER2-expression status using the fluorescein-labeled anti-HER2 antibody | 100 patients with histopathologic-confirmed UC post RC with bilateral lymphadenectomy | Clinical relevance of CTC HER2 expression in patients with non-metastatic UC | Significantly higher risks of disease recurrence, cancer-specific and overall mortality in CTC positive patients. No association of CTC HER2 status and clinicopathologic features | |
| De Santis et al. (103) | Platinum refractory UC | Prognosis | CTC enumeration in blood using CellSearch. Determination of CTC HER2-expression status using tumor phenotyping reagent HER2/neu | 70 patients with platinum refractory UC | Evaluation of CTC cut-off levels for drug screening and HER2 status on CTC in comparison to the primary tumor | CTCs detectable in 66% of patients. CTCs detected in all patients with HER2-positive primary tumors. HER2 positive CTC correlated with Bellmunt risk group 3 |
| Liu et al. (104) | MIBC and BC who underwent cystectomy and super-extended pelvic LN dissection | Prognosis/treatment selection | RNA extraction from urine and tissue samples and detection of miRNA-200b and miRNA-141 using qRT-PCR | 30 patients with MIBC and 78 others with BC who underwent cystectomy and super-extended pelvic LN dissection | Relation between miRNA-200b and miRNA-141, and the invasive ability and EMT of BC cells in vitro. Potential use of miRNA-200b and miRNA-141 to guide the extent of pelvic LN dissection during RC | MiRNA-200b and miRNA-141 essential in the invasive capability and EMT phenotype of BC, and predictive of LN metastasis |
| Powles et al. (105) | MIBC who underwent surgery | Treatment response | Plasma cfDNA extraction and multiplexed targeted PCR followed by amplicon-based sequencing | 581 patients with MIBC who underwent surgery | Outcomes of patient from a randomized phase III trial of adjuvant atezolizumab vs. observation in operable UC in correlation with their ctDNA status | Improved DFS and OS in ctDNA positive patients in the atezolizumab arm vs. the observation arm |
| Fanous et al. (106) | UC | Therapy | Profiling exosomal miRNAs from the resistant sublines of the human CUB III bladder carcinoma cell line using qRT-PCR | N/A | Role of specific miRNAs in chemoresistance of BC cells | Significant downregulation of miR-Let-7i-3p and upregulation of miR-21-5p → potential role in predicting treatment response and biomarkers for targeted therapy |
BC, bladder cancer; UC, urothelial carcinoma; LN, lymph node; aUC, advanced urothelial carcinoma; NMIBC, non-muscle invasive bladder cancer; MIBC, muscle invasive bladder cancer; cfDNA, cell free DNA; ctDNA, circulating tumor DNA; CTC, circulating tumor cell; LNMAT2, lymph node metastasis-associated transcript 2; EpCAM, epithelial cell adhesion molecule; FGFR3, fibroblast growth factor receptor 3; PIK3CA, phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha; ddPCR, droplet digital polymerase chain reaction; PCR, polymerase chain reaction; miRNA, microRNA; RT-PCR, reverse transcription polymerase chain reaction; PD-L1, programmed death ligand 1; HER2, human epidermal growth factor receptor 2; qRT-PCR, quantitative reverse transcription polymerase chain reaction; SBRT, stereotactic body radiation therapy; mUC, metastatic urothelial carcinoma; RC, radical cystectomy; TURB, transurethral resection of the bladder; BCG, Bacillus Calmette-Guerin; VEGF-C, vascular endothelial growth factor C; BRCA1, breast cancer gene 1; FFS, failure free survival; OS, overall survival; PFS, progression-free survival; RFS, recurrence-free survival; FDG, fluorodeoxyglucose; PET, positron emission tomography; CT, computed tomography; mTBI, molecular tumor burden index; AUC, area under the curve.
Role of liquid biopsy in BC detection and diagnosis
Many studies have investigated the role of different biomarkers in the diagnosis of BC. Urine tumor DNA are highly promising for the clinical detection of early stages BC (91,107-111). For instance, Dudley et al. reported high similarity for mutations between tumor and urine cfDNA after using a high‐throughput sequencing‐based hybrid capture method (111). Through this approach, it was demonstrated that pleckstrin homology domain containing S1 (PLEKHS1) promoter mutations, which are shed in urine, are among the most common somatic alterations in BC (111). Moreover, increased levels of ctDNA somatic variants were reported in MIBC, with concordance between ctDNA and tumor tissue (112,113). On the other hand, Welton et al. studied the exosomal expression of different protein biomarkers in cultured HT1376 BC cells (114). By using Western blotting and flow cytometry, they reported a significant expression of certain proteins like CD9, CD63 and CD81. They also detected 353 proteins in exosomes from BC by employing liquid chromatography. Then, after the elimination of any non-exosomal contaminant effect, the presence of 18 proteins including a6 integrin, β1 integrin CD73, CD10, CD36, mucin 1 (MUC1), basigin, and 5T4, was confirmed. This method was applied on urine from BC patients and control, and it revealed increased expression of CD36, CD44, 5T4, basigin, and CD73 among cancer patients when compared to control (114).
Recently, a group of researchers studied the DNA CNVs in the urine of patients with urothelial carcinoma (UC) (115). They developed an assay called Urine Exfoliated Cells Copy Number Aberration Detector (UroCAD) that analyzes urine samples, which they used to evaluate CNV in a total of 190 patients enrolled in an observational clinical trial in Shanghai. Among these patients, 126 had UC and 64 others were cancer free. The analysis revealed no significant presence of CNV in the cancer-free group. However, the investigators identified UC with a sensitivity of 82.5% and a specificity of 96.9%. Additionally, the same assay was used on 95 patients in a validation cohort, of whom 56 patients were diagnosed with UC. Despite the comparable specificity, UroCAD demonstrated significantly higher sensitivity when compared to urine cytology (80.4% vs. 33.9%). More importantly, in the 7 patients who had low-grade tumors located in the epithelial layer of the bladder, the sensitivity in detecting cancer was 71.4% for UroCAD compared to 0% with urine cytology. Accordingly, UroCAD was linked to tumor grade and tumor size with regard to sensitivity (115).
Perez et al. also shed the light on potential biomarkers that can be possibly used for diagnosing BC (116). After collecting urine samples of 5 BC patients and 6 non-BC patients before undergoing cystoscopy, they isolated exosomes, characterized them and used PCR to validate the array results on new samples. Among these, only exosomes from cancer patients expressed polypeptide N-acetylgalactosaminyltransferase 1 (GALNT1) and ceramide synthase 2 (CERS2) mRNAs. Although limited, such results presented potential RNA-associated to urinary vesicles that might be used for future non-invasive diagnostic purposes in BC (116). Studies also suggested that patients with high-grade MIBC demonstrated a urinary exosomal enrichment of specific long non-coding RNAs (lncRNAs) such as HOX transcript antisense RNA (HOTAIR) and metastasis associated lung adenocarcinoma transcript 1 (MALAT1), and other mRNAs such as SRY-box 2 (SOX2) and POU class 5 homeobox 1 (POU5F1) when compared to healthy patients (117). On that account, further studies and comparison in larger population of MIBC and healthy patients are needed to establish a significance for the use of these exosomal RNAs as biomarkers for diagnosing MIBC.
Currently, five urinary assays have been approved by the US FDA for initial detection of BC. These include nuclear matrix protein 22 (NMP22) kit, NMP22 BladderChek Test, bladder tumor antigen (BTA)-TRAK and BTA stat kits, Cell Search, and UroVysion (118). Although not widely used for diagnosing BC yet, it is shown promising that liquid biopsy will play a definitive role in the future diagnostic purposes of BC patients; and while it might not be enough to replace cystoscopy, it can still have an additive value and can probably limit the use of cystoscopy in the course of the disease.
Prognostic role of liquid biopsy in BC: progression, recurrence or survival
Studies suggest that liquid biopsy carries important prognostic values in BC patients. In fact, testing of ctDNA in BC has helped in detecting multiple alterations that could predict disease outcome. For example, mutations in both BRCA1 DNA repair associated gene and rapidly accelerated fibrosarcoma-1 (RAF-1) proto-oncogene were demonstrated as poor clinical indicators of BC patients (83). Moreover, ctDNA can be used as a predictor of disease recurrence in MIBC patients after initial curative treatment. Such role will help in early administration of systemic therapy before progression to metastatic disease (119). In fact, studies have proved increased levels of fibroblast growth factor receptor 3 (FGFR3) and phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha (PIK3CA) mutated DNA in urine and plasma samples of patients with progressive and metastastatic BC (91). Sundahl et al. studied the implication of ctDNA fraction in correlation with treatment response (84). In this phase 1 trial, the researchers reported that patients with BC who responded to treatment showed a rapid decline in ctDNA fraction, whereas non-responders had stable or even increased fractions (84). Additionally, in April 2021, Zhang et al. published the first study to use targeted sequencing of ctDNA for NMIBC patients (92). Based on an 861-gene panel, they compared somatic mutations in matched tumor DNA and plasma ctDNA. Zhang et al. reported that patients with T1 tumors showed higher detection rate of ctDNA and higher concordance rate of somatic mutations between ctDNA and tumor DNA than Ta patients, which suggest that ctDNA levels in blood correlate with disease invasiveness. It was also found that tumors of larger size also had increased levels of ctDNA in their plasma. More importantly, the study focused on studying the molecular tumor burden index (mTBI) in ctDNA, and it revealed that higher mTBI reflected shorter disease-free survival (DFS) in NMIBC patients who received transurethral resection of bladder followed by immunotherapy (92). Therefore, mTBI in ctDNA could be an independent prognostic indicator of DFS in these patients. Similarly, a study by Vandekerkhove et al. also demonstrated that high ctDNA levels independently predict aggressive disease in MIBC patients (85). It was also found that OS was significantly improved in patients with metastatic UC whose ctDNA fraction was below the first quartile (85).
As with ctDNA, many studies suggested a correlation between the presence of CTCs and the worse prognosis of patients with UC (89,93,94,101). A meta-analysis combining 30 studies and including more than 2,000 patients validated this disadvantageous impact of CTC on UC prognosis (120). Even the detection of a single CTC has established a negative impact on the outcomes of BC patients, unlike other solid malignancies that have a cut-off value of 3–5 CTC (25,89,94,121). In fact, multiple studies have demonstrated an association between the presence of CTC and a higher risk of disease recurrence and even mortality post radical cystectomy (89,93,94,101). It was also found that metastatic UC demonstrated increased number of CTC and higher detection rate compared to non-metastatic disease (88). For instance, detection rate varied between 26% and 91% in patients with lymph node metastasis, and between 33% and 100% in patient with visceral metastasis (86,88,102,103). Other than its relation to metastatic burden, the positivity of CTC demonstrated an association with imaging-proven metastatic disease on fluorodeoxyglucose (FDG)-PET-CT modality (90). Accordingly, CTC detection may be applied for monitoring disease progression and response to treatment.
Exosomes on the other hand are enriched from urine samples and may be potentially used as prognostic markers in BC. For example, lncRNA lymph node metastasis-associated transcript 2 (LNMAT2) serves as predictor for nodal dissemination with an area under the curve (AUC) of 0.881 (87). Lymphatic metastasis was also recognized with detection of miRNAs in urine of patients with BC, with a reported AUC of 0.704 for miRNA-141 and 0.647 for miRNA-200b (104). Other examples of the prognostic ability of miRNA include miR-214, which can be detected in the urine of NMIBC patients and signifies disease recurrence (95). Also, serum miR-152 and urinary miR-22-3p and miR-200a-3p are of indicators of recurrence risk in NMIBC (96,97). Few studies evaluated the use of miRNAs panels in the management of patients with NMIBC. Sapre et al. reported an AUC of 0.85 with urinary profiling of NMIBC patients using a panel of six miRNAs (98). This panel was found to be specifically applicable in identifying recurrence in burdensome disease, like T1 stage and high-volume cancer, with a sensitivity of 88.0% (98). As such, this panel can help in the disease surveillance process and limit the use of invasive cystoscopies. Another study by Jiang et al. presented a panel of 4 miRNAs that predicted MIBC development and progression with AUC values significantly higher than those of cytology and BC grade (100).
Therapeutic use of liquid biopsy in BC
The use of liquid biopsy has been emerging lately as a guide for targeted therapy in solid tumors. Over the past years, multiple studies have been focusing on this specific concept for its potential value and importance in BC patients. Grivas et al. determined that molecular alterations within cfDNA were widely detected in patients with advanced UC (aUC) (83). In fact, detection of breast cancer gene 1 (BRCA1) and RAF1 alterations was specifically correlated to shortened OS and failure free survival (FFS). Also, given the pre-existent evidence of susceptibility to PARP inhibition in tumors with homologous DNA repair defects, it was suggested that targeted therapy might be employed in aUC carrying similar mutations. Likewise, a potential role of RAF kinase inhibitors was proposed in the treatment of aUC patients (83,122). Hence, cfDNA profiling could direct physicians toward the use of certain therapeutic strategies. Powles et al. evaluated outcomes in 581 patients who underwent surgery for UC, followed by atezolizumab vs. observation (105). Among the different study groups, ctDNA testing was done at different time-points. Through this study, it was shown that patients with positive ctDNA demonstrated an improved DFS and OS after immunotherapy, compared to those in the observation arm. However, for those who were negative for ctDNA, no difference was noted in DFS or OS in both treatment arms (105). Subsequently, ctDNA may be potentially used as a predictor for response to atezolizumab.
The gold standard treatment for NMIBC is intravesical Bacillus Calmette-Guerin (BCG) immunotherapy (123). However, not all NMIBC are responsive to BCG, and sensitivity was found to be altered by programmed death-ligand 1 (PD-L1) expression. Therefore, the US FDA approved anti-programmed death-1 (PD-1) monotherapy as treatment of BCG-unresponsive NMIBC patients (123,124). Recently, Nicolazzo et al. explored the possible role of liquid biopsy in the stratification of NMIBC according to their response to BCG (99). The researchers investigated CTC expression of PD-L1 in 20 CTC-positive NMIBC patients. CTC isolation was also compared between two methods: the antigen-dependent FDA-approved CellSearch system, and ScreenCell, a size-based CTC isolation method. After CTC isolation, patients were enrolled for adjuvant BCG therapy, and followed up for a median period of 18 months. Through the follow-up period, it was found that among the 20 enrolled patients, 12 patients were BCG-unresponsive, while 8 were BCG-responsive. Moreover, of the BCG-unresponsive group, 11 out of 12 patients demonstrated PD-L1-positive CTCs (92%), while CTCs from all patients in the response group were PD-L1 negative (99). As such, the use of CTC to determine PD-L1 status in NMIBC could be implemented in the selection of patients for BCG treatment or PD-L1 targeted immunotherapy.
Additionally, a study by Fanous et al. investigated exosomal miRNAs that could possibly affect the sensitivity of BC to cisplatin, gemcitabine, and cisplatin/gemcitabine (106). Over a period of 6 months, resistant BC cells were subject to gradually increased doses of cisplatin, gemcitabine, or a combination of cisplatin and gemcitabine. After profound analysis, it was found that for each type of therapy, resistant cells expressed specific exosomal miRNA profiles. Moreover, results showed that, among the profiled miRNAs, the most significantly downregulated was miR-Let-7i-3p, whereas miR-21-5p was the most significantly upregulated (106). Consequently, with further validation, miR-Let-7i-3p and miR-21-5p could potentially serve as biomarkers for treatment response monitoring and possible targeted therapy.
Conclusions
Liquid biopsy has a promising role in detecting BC and PC, predicting prognosis, steering treatment selection and monitoring for response. Multiple detection technics of the different biomarkers exist, and the assays used need to be standardized and validated, since the interpretation of results depends on the detection method used. Table 6 summarizes the main diagnostic and prognostic methods available for PC and BC. Although the use of liquid biopsy in localized and mHSPC is still low yield and limited, a substantial amount of evidence exists on its utility in mCRPC. The role of CTC count as a prognostic biomarker is established across multiple large studies, and it was found to correlate with response to chemotherapy. The detection of AR-V7 has also a prognostic and predictive value in determining treatment with taxanes vs. novel antiandrogens. The use of blood-based tests to detect HRR gene alteration is approved to guide treatment selection, and identify patients who would benefit from PARP inhibitors. While the use of liquid biopsy has already been approved in the diagnosis and surveillance of BC, the main focus and challenge remains in directing this revolutionary modality toward a new therapeutic horizon, that could establish a solid ground for patient selection for adjuvant and targeted therapy in those with BC. These biomarkers have to be evaluated in larger prospective trials, in order to establish their clinical utility and cost-effectiveness in guiding therapeutic decisions for individual patients.
Table 6
| Application | Advantages | Disadvantages | Test performance | Reference |
|---|---|---|---|---|
| TRUS biopsy | ||||
| PC diagnosis | Standard of practice, accessible to patients | Invasive, inadequate sampling of tumor heterogeneity | 48% sensitivity and 96% specificity | (125) |
| PSA | ||||
| PC diagnosis, prognosis monitoring | Less invasive than tissue biopsy | Poor discrimination of malignant and benign diseases | 21% sensitivity and 91% specificity (cut-off of 4.0 ng/mL) | (126) |
| Cystoscopy | ||||
| BC diagnosis | Standard of practice, direct tumor visualization, accessible to patients | Invasive, inadequate sampling of small flat tumors | 62–84% sensitivity and 43–98% specificity | (127) |
| CfDNA | ||||
| PC and BC diagnosis, prognosis and monitoring | Less invasive than tissue biopsy, real-time monitoring of disease, reflects tumor heterogeneity | Processing is technologically complex and not readily available, methods need standardization | PC diagnosis: 80% sensitivity and 82% specificity. BC diagnosis: 93.5% sensitivity and 95.8% specificity. CfDNA concentration independent predictor of OS in PC (HR: 0.34, P=0.032) and BC (HR: 3.59) | (1,39,68,85,128) |
| CTC count | ||||
| BC diagnosis, PC and BC prognosis and monitoring | Less invasive than tissue biopsy, real-time monitoring of disease, reflects tumor heterogeneity | Processing is technologically complex and not readily available, methods need standardization | 35% sensitivity and 97 specificity (BC diagnosis). CTC count ≥5 cells/7.5 mL correlates with lower OS in CRPC (HR: 3.23, P=0.0028) and BC (HR: 3.98, P<0.001) | (37,120) |
PC, prostate cancer; BC, bladder cancer; TRUS, transrectal ultrasonography; PSA, prostate specific antigen; cfDNA, cell free DNA; OS, overall survival; HR, hazard ratio; CTC, circulating tumor cell; CRPC, castrate resistant prostate cancer.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Elie Rassy, Nicholas Pavlidis, Stergios Boussios, and Joseph Kattan) for the series “Precision Oncology in Urogenital and Gynecological Tumors” published in Precision Cancer Medicine. The article has undergone external peer review.
Reporting Checklist: The authors have completed the Narrative Review reporting checklist. Available https://pcm.amegroups.com/article/view/10.21037/pcm-22-1/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://pcm.amegroups.com/article/view/10.21037/pcm-22-1/coif). The series “Precision Oncology in Urogenital and Gynecological Tumors” was commissioned by the editorial office without any funding or sponsorship. DM reports institutional research support from Astellas and BMS, as well as honoraria for lectures from: Astellas, Janssen, BMS, Bayer, MSD, Ipsen. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Di Meo A, Bartlett J, Cheng Y, et al. Liquid biopsy: a step forward towards precision medicine in urologic malignancies. Mol Cancer 2017;16:80. [Crossref] [PubMed]
- Diamandis M, White NM, Yousef GM. Personalized medicine: marking a new epoch in cancer patient management. Mol Cancer Res 2010;8:1175-87. [Crossref] [PubMed]
- Ignatiadis M, Sledge GW, Jeffrey SS. Liquid biopsy enters the clinic - implementation issues and future challenges. Nat Rev Clin Oncol 2021;18:297-312. [Crossref] [PubMed]
- Fernández-Lázaro D, García Hernández JL, García AC, et al. Liquid Biopsy as Novel Tool in Precision Medicine: Origins, Properties, Identification and Clinical Perspective of Cancer's Biomarkers. Diagnostics (Basel) 2020;10:215. [Crossref] [PubMed]
- Cieślikowski WA, Antczak A, Nowicki M, et al. Clinical Relevance of Circulating Tumor Cells in Prostate Cancer Management. Biomedicines 2021;9:1179. [Crossref] [PubMed]
- Puche-Sanz I, Rodríguez-Martínez A, Garrido-Navas MC, et al. Liquid biopsy and prostate cancer. Current evidence applied to clinical practice. Actas Urol Esp 2020;44:139-47. (Engl Ed). [Crossref] [PubMed]
- Miller MC, Doyle GV, Terstappen LW. Significance of Circulating Tumor Cells Detected by the CellSearch System in Patients with Metastatic Breast Colorectal and Prostate Cancer. J Oncol 2010;2010:617421. [Crossref] [PubMed]
- Pantel K, Hille C, Scher HI. Circulating Tumor Cells in Prostate Cancer: From Discovery to Clinical Utility. Clin Chem 2019;65:87-99. [Crossref] [PubMed]
- Danila DC, Fleisher M, Scher HI. Circulating tumor cells as biomarkers in prostate cancer. Clin Cancer Res 2011;17:3903-12. [Crossref] [PubMed]
- Alix-Panabières C. EPISPOT assay: detection of viable DTCs/CTCs in solid tumor patients. Recent Results Cancer Res 2012;195:69-76. [Crossref] [PubMed]
- Zainfeld D, Goldkorn A. Liquid Biopsy in Prostate Cancer: Circulating Tumor Cells and Beyond. Cancer Treat Res 2018;175:87-104. [Crossref] [PubMed]
- Ocansey DKW, Zhang L, Wang Y, et al. Exosome-mediated effects and applications in inflammatory bowel disease. Biol Rev Camb Philos Soc 2020;95:1287-307. [Crossref] [PubMed]
- Skog J, Würdinger T, van Rijn S, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol 2008;10:1470-6. [Crossref] [PubMed]
- Martins VR, Dias MS, Hainaut P. Tumor-cell-derived microvesicles as carriers of molecular information in cancer. Curr Opin Oncol 2013;25:66-75. [Crossref] [PubMed]
- Hessvik NP, Llorente A. Current knowledge on exosome biogenesis and release. Cell Mol Life Sci 2018;75:193-208. [Crossref] [PubMed]
- Franzen CA, Blackwell RH, Foreman KE, et al. Urinary Exosomes: The Potential for Biomarker Utility, Intercellular Signaling and Therapeutics in Urological Malignancy. J Urol 2016;195:1331-9. [Crossref] [PubMed]
- Blackwell RH, Franzen CA, Flanigan RC, et al. The untapped potential of urine shed bladder cancer exosomes: biomarkers, signaling, and therapeutics. Bladder 2014;1:e7. [Crossref]
- Salvi S, Bandini E, Fabbri F. Urinary Exosomes in Prostate Cancer. Methods Mol Biol 2021;2292:115-20. [Crossref] [PubMed]
- Knezevic D, Goddard AD, Natraj N, et al. Analytical validation of the Oncotype DX prostate cancer assay - a clinical RT-PCR assay optimized for prostate needle biopsies. BMC Genomics 2013;14:690. [Crossref] [PubMed]
- Sartori DA, Chan DW. Biomarkers in prostate cancer: what's new? Curr Opin Oncol 2014;26:259-64. [Crossref] [PubMed]
- Constâncio V, Nunes SP, Henrique R, et al. DNA Methylation-Based Testing in Liquid Biopsies as Detection and Prognostic Biomarkers for the Four Major Cancer Types. Cells 2020;9:624. [Crossref] [PubMed]
- Sánchez BE, Aguayo A, Martínez B, et al. Using Genetic and Epigenetic Markers to Improve Differential Diagnosis of Prostate Cancer and Benign Prostatic Hyperplasia by Noninvasive Methods in Mexican Patients. Clin Genitourin Cancer 2018;16:e867-77. [Crossref] [PubMed]
- McKiernan J, Donovan MJ, O'Neill V, et al. A Novel Urine Exosome Gene Expression Assay to Predict High-grade Prostate Cancer at Initial Biopsy. JAMA Oncol 2016;2:882-9. [Crossref] [PubMed]
- Danila DC, Heller G, Gignac GA, et al. Circulating tumor cell number and prognosis in progressive castration-resistant prostate cancer. Clin Cancer Res 2007;13:7053-8. [Crossref] [PubMed]
- de Bono JS, Scher HI, Montgomery RB, et al. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res 2008;14:6302-9. [Crossref] [PubMed]
- Scher HI, Jia X, de Bono JS, et al. Circulating tumour cells as prognostic markers in progressive, castration-resistant prostate cancer: a reanalysis of IMMC38 trial data. Lancet Oncol 2009;10:233-9. [Crossref] [PubMed]
- Huang X, Yuan T, Liang M, et al. Exosomal miR-1290 and miR-375 as prognostic markers in castration-resistant prostate cancer. Eur Urol 2015;67:33-41. [Crossref] [PubMed]
- Del Re M, Biasco E, Crucitta S, et al. The Detection of Androgen Receptor Splice Variant 7 in Plasma-derived Exosomal RNA Strongly Predicts Resistance to Hormonal Therapy in Metastatic Prostate Cancer Patients. Eur Urol 2017;71:680-7. [Crossref] [PubMed]
- De Laere B, Oeyen S, Mayrhofer M, et al. TP53 Outperforms Other Androgen Receptor Biomarkers to Predict Abiraterone or Enzalutamide Outcome in Metastatic Castration-Resistant Prostate Cancer. Clin Cancer Res 2019;25:1766-73. [Crossref] [PubMed]
- Mehra N, Dolling D, Sumanasuriya S, et al. Plasma Cell-free DNA Concentration and Outcomes from Taxane Therapy in Metastatic Castration-resistant Prostate Cancer from Two Phase III Trials (FIRSTANA and PROSELICA). Eur Urol 2018;74:283-91. [Crossref] [PubMed]
- Goodall J, Mateo J, Yuan W, et al. Circulating Cell-Free DNA to Guide Prostate Cancer Treatment with PARP Inhibition. Cancer Discov 2017;7:1006-17. [Crossref] [PubMed]
- Torquato S, Pallavajjala A, Goldstein A, et al. Genetic Alterations Detected in Cell-Free DNA Are Associated With Enzalutamide and Abiraterone Resistance in Castration-Resistant Prostate Cancer. JCO Precis Oncol 2019;3:PO.18.00227.
- Okegawa T, Itaya N, Hara H, et al. Circulating tumor cells as a biomarker predictive of sensitivity to docetaxel chemotherapy in patients with castration-resistant prostate cancer. Anticancer Res 2014;34:6705-10. [PubMed]
- Vogelzang NJ, Fizazi K, Burke JM, et al. Circulating Tumor Cells in a Phase 3 Study of Docetaxel and Prednisone with or without Lenalidomide in Metastatic Castration-resistant Prostate Cancer. Eur Urol 2017;71:168-71. [Crossref] [PubMed]
- Goldkorn A, Ely B, Quinn DI, et al. Circulating tumor cell counts are prognostic of overall survival in SWOG S0421: a phase III trial of docetaxel with or without atrasentan for metastatic castration-resistant prostate cancer. J Clin Oncol 2014;32:1136-42. [Crossref] [PubMed]
- Heller G, McCormack R, Kheoh T, et al. Circulating Tumor Cell Number as a Response Measure of Prolonged Survival for Metastatic Castration-Resistant Prostate Cancer: A Comparison With Prostate-Specific Antigen Across Five Randomized Phase III Clinical Trials. J Clin Oncol 2018;36:572-80. [Crossref] [PubMed]
- Lorente D, Olmos D, Mateo J, et al. Circulating tumour cell increase as a biomarker of disease progression in metastatic castration-resistant prostate cancer patients with low baseline CTC counts. Ann Oncol 2018;29:1554-60. [Crossref] [PubMed]
- Scher HI, Heller G, Molina A, et al. Circulating tumor cell biomarker panel as an individual-level surrogate for survival in metastatic castration-resistant prostate cancer. J Clin Oncol 2015;33:1348-55. [Crossref] [PubMed]
- Kienel A, Porres D, Heidenreich A, et al. cfDNA as a Prognostic Marker of Response to Taxane Based Chemotherapy in Patients with Prostate Cancer. J Urol 2015;194:966-71. [Crossref] [PubMed]
- Annala M, Vandekerkhove G, Khalaf D, et al. Circulating Tumor DNA Genomics Correlate with Resistance to Abiraterone and Enzalutamide in Prostate Cancer. Cancer Discov 2018;8:444-57. [Crossref] [PubMed]
- Conteduca V, Wetterskog D, Sharabiani MTA, et al. Androgen receptor gene status in plasma DNA associates with worse outcome on enzalutamide or abiraterone for castration-resistant prostate cancer: a multi-institution correlative biomarker study. Ann Oncol 2017;28:1508-16. [Crossref] [PubMed]
- Antonarakis ES, Lu C, Wang H, et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med 2014;371:1028-38. [Crossref] [PubMed]
- Antonarakis ES, Lu C, Luber B, et al. Clinical Significance of Androgen Receptor Splice Variant-7 mRNA Detection in Circulating Tumor Cells of Men With Metastatic Castration-Resistant Prostate Cancer Treated With First- and Second-Line Abiraterone and Enzalutamide. J Clin Oncol 2017;35:2149-56. [Crossref] [PubMed]
- Scher HI, Graf RP, Schreiber NA, et al. Assessment of the Validity of Nuclear-Localized Androgen Receptor Splice Variant 7 in Circulating Tumor Cells as a Predictive Biomarker for Castration-Resistant Prostate Cancer. JAMA Oncol 2018;4:1179-86. [Crossref] [PubMed]
- Darshan MS, Loftus MS, Thadani-Mulero M, et al. Taxane-induced blockade to nuclear accumulation of the androgen receptor predicts clinical responses in metastatic prostate cancer. Cancer Res 2011;71:6019-29. [Crossref] [PubMed]
- Salvi S, Casadio V, Conteduca V, et al. Circulating cell-free AR and CYP17A1 copy number variations may associate with outcome of metastatic castration-resistant prostate cancer patients treated with abiraterone. Br J Cancer 2015;112:1717-24. [Crossref] [PubMed]
- Armstrong AJ, Halabi S, Luo J, et al. Prospective Multicenter Validation of Androgen Receptor Splice Variant 7 and Hormone Therapy Resistance in High-Risk Castration-Resistant Prostate Cancer: The PROPHECY Study. J Clin Oncol 2019;37:1120-9. [Crossref] [PubMed]
- Goodman OB Jr, Symanowski JT, Loudyi A, et al. Circulating tumor cells as a predictive biomarker in patients with hormone-sensitive prostate cancer. Clin Genitourin Cancer 2011;9:31-8. [Crossref] [PubMed]
- Josefsson A, Damber JE, Welén K. AR-V7 expression in circulating tumor cells as a potential prognostic marker in metastatic hormone-sensitive prostate cancer. Acta Oncol 2019;58:1660-4. [Crossref] [PubMed]
- Yang YJ, Kong YY, Li GX, et al. Phenotypes of circulating tumour cells predict time to castration resistance in metastatic castration-sensitive prostate cancer. BJU Int 2019;124:258-67. [Crossref] [PubMed]
- Thalgott M, Rack B, Horn T, et al. Detection of Circulating Tumor Cells in Locally Advanced High-risk Prostate Cancer During Neoadjuvant Chemotherapy and Radical Prostatectomy. Anticancer Res 2015;35:5679-85. [PubMed]
- Meyer CP, Pantel K, Tennstedt P, et al. Limited prognostic value of preoperative circulating tumor cells for early biochemical recurrence in patients with localized prostate cancer. Urol Oncol 2016;34:235.e11-6. [Crossref] [PubMed]
- Loh J, Jovanovic L, Lehman M, et al. Circulating tumor cell detection in high-risk non-metastatic prostate cancer. J Cancer Res Clin Oncol 2014;140:2157-62. [Crossref] [PubMed]
- Davis JW, Nakanishi H, Kumar VS, et al. Circulating tumor cells in peripheral blood samples from patients with increased serum prostate specific antigen: initial results in early prostate cancer. J Urol 2008;179:2187-91; discussion 2191. [Crossref] [PubMed]
- Aragon-Ching JB, Siegel RS, Frazier H 2nd, et al. Circulating Tumor Cells in Biochemical Recurrence of Prostate Cancer. Clin Genitourin Cancer 2015;13:e341-5. [Crossref] [PubMed]
- Todenhöfer T, Park ES, Duffy S, et al. Microfluidic enrichment of circulating tumor cells in patients with clinically localized prostate cancer. Urol Oncol 2016;34:483.e9-483.e16. [Crossref] [PubMed]
- Friedlander TW, Welty C, Anantharaman A, et al. Identification and Characterization of Circulating Tumor Cells in Men Who have Undergone Prostatectomy for Clinically Localized, High Risk Prostate Cancer. J Urol 2019;202:732-41. [Crossref] [PubMed]
- Ellinger J, Haan K, Heukamp LC, et al. CpG island hypermethylation in cell-free serum DNA identifies patients with localized prostate cancer. Prostate 2008;68:42-9. [Crossref] [PubMed]
- Hendriks RJ, van Oort IM, Schalken JA. Blood-based and urinary prostate cancer biomarkers: a review and comparison of novel biomarkers for detection and treatment decisions. Prostate Cancer Prostatic Dis 2017;20:12-9. [Crossref] [PubMed]
- Boerrigter E, Groen LN, Van Erp NP, et al. Clinical utility of emerging biomarkers in prostate cancer liquid biopsies. Expert Rev Mol Diagn 2020;20:219-30. [Crossref] [PubMed]
- Danila DC, Pantel K, Fleisher M, et al. Circulating tumors cells as biomarkers: progress toward biomarker qualification. Cancer J 2011;17:438-50. [Crossref] [PubMed]
- van Dessel LF, Martens JWM, Lolkema MP. Fundamentals of liquid biopsies in metastatic prostate cancer: from characterization to stratification. Curr Opin Oncol 2020;32:527-34. [Crossref] [PubMed]
- Kwee S, Song MA, Cheng I, et al. Measurement of circulating cell-free DNA in relation to 18F-fluorocholine PET/CT imaging in chemotherapy-treated advanced prostate cancer. Clin Transl Sci 2012;5:65-70. [Crossref] [PubMed]
- National Comprehensive Cancer Network. Prostate Cancer (Version 1.2022). 2022. Available online: https://www.nccn.org/professionals/physician_gls/pdf/prostate.pdf
- FDA. FDA approves olaparib for HRR gene-mutated metastatic castration-resistant prostate cancer. 2020. Available online: https://www.fda.gov/drugs/resources-information-approved-drugs/fda-approves-olaparib-hrr-gene-mutated-metastatic-castration-resistant-prostate-cancer
- Kuske A, Gorges TM, Tennstedt P, et al. Improved detection of circulating tumor cells in non-metastatic high-risk prostate cancer patients. Sci Rep 2016;6:39736. [Crossref] [PubMed]
- Cieślikowski WA, Budna-Tukan J, Świerczewska M, et al. Circulating Tumor Cells as a Marker of Disseminated Disease in Patients with Newly Diagnosed High-Risk Prostate Cancer. Cancers (Basel) 2020;12:160. [Crossref] [PubMed]
- Vandekerkhove G, Struss WJ, Annala M, et al. Circulating Tumor DNA Abundance and Potential Utility in De Novo Metastatic Prostate Cancer. Eur Urol 2019;75:667-75. [Crossref] [PubMed]
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Sanli O, Dobruch J, Knowles MA, et al. Bladder cancer. Nat Rev Dis Primers 2017;3:17022. [Crossref] [PubMed]
- National Comprehensive Cancer Network. Bladder Cancer (Version: 5.2021). 2021. Available online: https://www.nccn.org/professionals/physician_gls/pdf/bladder.pdf
- Sylvester RJ, van der Meijden AP, Oosterlinck W, et al. Predicting recurrence and progression in individual patients with stage Ta T1 bladder cancer using EORTC risk tables: a combined analysis of 2596 patients from seven EORTC trials. Eur Urol 2006;49:466-5; discussion 475-7. [Crossref] [PubMed]
- Su H, Jiang H, Tao T, et al. Hope and challenge: Precision medicine in bladder cancer. Cancer Med 2019;8:1806-16. [Crossref] [PubMed]
- Lawrence MS, Stojanov P, Polak P, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 2013;499:214-8. [Crossref] [PubMed]
- Heide T, Maurer A, Eipel M, et al. Multiregion human bladder cancer sequencing reveals tumour evolution, bladder cancer phenotypes and implications for targeted therapy. J Pathol 2019;248:230-42. [Crossref] [PubMed]
- Thomsen MBH, Nordentoft I, Lamy P, et al. Comprehensive multiregional analysis of molecular heterogeneity in bladder cancer. Sci Rep 2017;7:11702. [Crossref] [PubMed]
- Kamoun A, de Reyniès A, Allory Y, et al. A Consensus Molecular Classification of Muscle-invasive Bladder Cancer. Eur Urol 2020;77:420-33. [Crossref] [PubMed]
- Wan JCM, Massie C, Garcia-Corbacho J, et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer 2017;17:223-38. [Crossref] [PubMed]
- Zeng C, Stroup EK, Zhang Z, et al. Towards precision medicine: advances in 5-hydroxymethylcytosine cancer biomarker discovery in liquid biopsy. Cancer Commun (Lond) 2019;39:12. [Crossref] [PubMed]
- Cheng T, Roth B, Choi W, et al. Fibroblast growth factor receptors-1 and -3 play distinct roles in the regulation of bladder cancer growth and metastasis: implications for therapeutic targeting. PLoS One 2013;8:e57284. [Crossref] [PubMed]
- Georgantzoglou N, Pergaris A, Masaoutis C, et al. Extracellular Vesicles as Biomarkers Carriers in Bladder Cancer: Diagnosis, Surveillance, and Treatment. Int J Mol Sci 2021;22:2744. [Crossref] [PubMed]
- Kouba E, Lopez-Beltran A, Montironi R, et al. Liquid biopsy in the clinical management of bladder cancer: current status and future developments. Expert Rev Mol Diagn 2020;20:255-64. [Crossref] [PubMed]
- Grivas P, Lalani AA, Pond GR, et al. Circulating Tumor DNA Alterations in Advanced Urothelial Carcinoma and Association with Clinical Outcomes: A Pilot Study. Eur Urol Oncol 2020;3:695-9. [Crossref] [PubMed]
- Sundahl N, Vandekerkhove G, Decaestecker K, et al. Randomized Phase 1 Trial of Pembrolizumab with Sequential Versus Concomitant Stereotactic Body Radiotherapy in Metastatic Urothelial Carcinoma. Eur Urol 2019;75:707-11. [Crossref] [PubMed]
- Vandekerkhove G, Lavoie JM, Annala M, et al. Plasma ctDNA is a tumor tissue surrogate and enables clinical-genomic stratification of metastatic bladder cancer. Nat Commun 2021;12:184. [Crossref] [PubMed]
- Gallagher DJ, Milowsky MI, Ishill N, et al. Detection of circulating tumor cells in patients with urothelial cancer. Ann Oncol 2009;20:305-8. [Crossref] [PubMed]
- Chen C, Luo Y, He W, et al. Exosomal long noncoding RNA LNMAT2 promotes lymphatic metastasis in bladder cancer. J Clin Invest 2020;130:404-21. [Crossref] [PubMed]
- Okegawa T, Hayashi K, Hara H, et al. Immunomagnetic quantification of circulating tumor cells in patients with urothelial cancer. Int J Urol 2010;17:254-8. [Crossref] [PubMed]
- Rink M, Chun FK, Minner S, et al. Detection of circulating tumour cells in peripheral blood of patients with advanced non-metastatic bladder cancer. BJU Int 2011;107:1668-75. [Crossref] [PubMed]
- Abrahamsson J, Aaltonen K, Engilbertsson H, et al. Circulating tumor cells in patients with advanced urothelial carcinoma of the bladder: Association with tumor stage, lymph node metastases, FDG-PET findings, and survival. Urol Oncol 2017;35:606.e9-606.e16. [Crossref] [PubMed]
- Christensen E, Birkenkamp-Demtröder K, Nordentoft I, et al. Liquid Biopsy Analysis of FGFR3 and PIK3CA Hotspot Mutations for Disease Surveillance in Bladder Cancer. Eur Urol 2017;71:961-9. [Crossref] [PubMed]
- Zhang J, Dai D, Tian J, et al. Circulating Tumor DNA Analyses Predict Disease Recurrence in Non-Muscle-Invasive Bladder Cancer. Front Oncol 2021;11:657483. [Crossref] [PubMed]
- Guzzo TJ, McNeil BK, Bivalacqua TJ, et al. The presence of circulating tumor cells does not predict extravesical disease in bladder cancer patients prior to radical cystectomy. Urol Oncol 2012;30:44-8. [Crossref] [PubMed]
- Soave A, Riethdorf S, Dahlem R, et al. A nonrandomized, prospective, clinical study on the impact of circulating tumor cells on outcomes of urothelial carcinoma of the bladder patients treated with radical cystectomy with or without adjuvant chemotherapy. Int J Cancer 2017;140:381-9. [Crossref] [PubMed]
- Kim SM, Kang HW, Kim WT, et al. Cell-Free microRNA-214 From Urine as a Biomarker for Non-Muscle-Invasive Bladder Cancer. Korean J Urol 2013;54:791-6. [Crossref] [PubMed]
- Jiang X, Du L, Wang L, et al. Serum microRNA expression signatures identified from genome-wide microRNA profiling serve as novel noninvasive biomarkers for diagnosis and recurrence of bladder cancer. Int J Cancer 2015;136:854-62. [Crossref] [PubMed]
- Du L, Jiang X, Duan W, et al. Cell-free microRNA expression signatures in urine serve as novel noninvasive biomarkers for diagnosis and recurrence prediction of bladder cancer. Oncotarget 2017;8:40832-42. [Crossref] [PubMed]
- Sapre N, Macintyre G, Clarkson M, et al. A urinary microRNA signature can predict the presence of bladder urothelial carcinoma in patients undergoing surveillance. Br J Cancer 2016;114:454-62. [Crossref] [PubMed]
- Nicolazzo C, de Berardinis E, Gazzaniga P. Liquid Biopsy for Predicting Bacillus Calmette-Guérin Unresponsiveness in Non-muscle-invasive Bladder Cancer. Eur Urol Oncol 2021;4:124-5. [Crossref] [PubMed]
- Jiang X, Du L, Duan W, et al. Serum microRNA expression signatures as novel noninvasive biomarkers for prediction and prognosis of muscle-invasive bladder cancer. Oncotarget 2016;7:36733-42. [Crossref] [PubMed]
- Rink M, Soave A, Chun FK, et al. 876 Preoperative detection of a single circulating tumour cell is independently associated with inferior outcome in patients with urothelial carcinoma of the bladder. J Urol 2012;187:e356-7. [Crossref]
- Rink M, Chun FK, Dahlem R, et al. Prognostic role and HER2 expression of circulating tumor cells in peripheral blood of patients prior to radical cystectomy: a prospective study. Eur Urol 2012;61:810-7. [Crossref] [PubMed]
- De Santis M, Bauernhofer T, Stoiber F, et al. Circulating tumor cells (CTC) and HER-2 status on CTC and primary tumor in urothelial cancer (UC) patients refractory to platinum based chemotherapy. J Clin Oncol 2016;34:abstr 4520.
- Liu W, Qi L, Lv H, et al. MiRNA-141 and miRNA-200b are closely related to invasive ability and considered as decision-making biomarkers for the extent of PLND during cystectomy. BMC Cancer 2015;15:92. [Crossref] [PubMed]
- Powles T, Assaf ZJ, Davarpanah N, et al. ctDNA guiding adjuvant immunotherapy in urothelial carcinoma. Nature 2021;595:432-7. [Crossref] [PubMed]
- Fanous H, Sullivan T, Rieger-Christ K. MP88-15 Distinct exosomal mirna profiles in chemoresistant bladder carcinoma cell lines. J Urol 2017;197:e1179-80. [Crossref]
- Springer SU, Chen CH, Rodriguez Pena MDC, et al. Non-invasive detection of urothelial cancer through the analysis of driver gene mutations and aneuploidy. Elife 2018;7:32143. [Crossref]
- Ward DG, Baxter L, Gordon NS, et al. Multiplex PCR and Next Generation Sequencing for the Non-Invasive Detection of Bladder Cancer. PLoS One 2016;11:e0149756. [Crossref] [PubMed]
- Hirotsu Y, Yokoyama H, Amemiya K, et al. Genomic profile of urine has high diagnostic sensitivity compared to cytology in non-invasive urothelial bladder cancer. Cancer Sci 2019;110:3235-43. [Crossref] [PubMed]
- Hayashi Y, Fujita K, Matsuzaki K, et al. Diagnostic potential of TERT promoter and FGFR3 mutations in urinary cell-free DNA in upper tract urothelial carcinoma. Cancer Sci 2019;110:1771-9. [Crossref] [PubMed]
- Dudley JC, Schroers-Martin J, Lazzareschi DV, et al. Detection and Surveillance of Bladder Cancer Using Urine Tumor DNA. Cancer Discov 2019;9:500-9. [Crossref] [PubMed]
- Birkenkamp-Demtröder K, Nordentoft I, Christensen E, et al. Genomic Alterations in Liquid Biopsies from Patients with Bladder Cancer. Eur Urol 2016;70:75-82. [Crossref] [PubMed]
- Grivas P, Nagy RJ, Pond GR, et al. Circulating tumor (ct)-DNA alterations in urothelial/bladder cancer (UC/BC): Updates on a dynamic genomic landscape. J Clin Oncol 2017;35:abstr 4534.
- Welton JL, Khanna S, Giles PJ, et al. Proteomics analysis of bladder cancer exosomes. Mol Cell Proteomics 2010;9:1324-38. [Crossref] [PubMed]
- Zeng S, Ying Y, Xing N, et al. Noninvasive Detection of Urothelial Carcinoma by Cost-effective Low-coverage Whole-genome Sequencing from Urine-Exfoliated Cell DNA. Clin Cancer Res 2020;26:5646-54. [Crossref] [PubMed]
- Perez A, Loizaga A, Arceo R, et al. A Pilot Study on the Potential of RNA-Associated to Urinary Vesicles as a Suitable Non-Invasive Source for Diagnostic Purposes in Bladder Cancer. Cancers (Basel) 2014;6:179-92. [Crossref] [PubMed]
- Berrondo C, Flax J, Kucherov V, et al. Expression of the Long Non-Coding RNA HOTAIR Correlates with Disease Progression in Bladder Cancer and Is Contained in Bladder Cancer Patient Urinary Exosomes. PLoS One 2016;11:e0147236. [Crossref] [PubMed]
- Sugeeta SS, Sharma A, Ng K, et al. Biomarkers in Bladder Cancer Surveillance. Front Surg 2021;8:735868. [Crossref] [PubMed]
- Todenhöfer T, Struss WJ, Seiler R, et al. Liquid Biopsy-Analysis of Circulating Tumor DNA (ctDNA) in Bladder Cancer. Bladder Cancer 2018;4:19-29. [Crossref] [PubMed]
- Zhang Z, Fan W, Deng Q, et al. The prognostic and diagnostic value of circulating tumor cells in bladder cancer and upper tract urothelial carcinoma: a meta-analysis of 30 published studies. Oncotarget 2017;8:59527-38. [Crossref] [PubMed]
- Cohen SJ, Punt CJ, Iannotti N, et al. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J Clin Oncol 2008;26:3213-21. [Crossref] [PubMed]
- Kaufman B, Shapira-Frommer R, Schmutzler RK, et al. Olaparib monotherapy in patients with advanced cancer and a germline BRCA1/2 mutation. J Clin Oncol 2015;33:244-50. [Crossref] [PubMed]
- Tse J, Singla N, Ghandour R, et al. Current advances in BCG-unresponsive non-muscle invasive bladder cancer. Expert Opin Investig Drugs 2019;28:757-70. [Crossref] [PubMed]
- Grimm MO, Bex A, De Santis M, et al. Safe Use of Immune Checkpoint Inhibitors in the Multidisciplinary Management of Urological Cancer: The European Association of Urology Position in 2019. Eur Urol 2019;76:368-80. [Crossref] [PubMed]
- Ahmed HU, El-Shater Bosaily A, Brown LC, et al. Diagnostic accuracy of multi-parametric MRI and TRUS biopsy in prostate cancer (PROMIS): a paired validating confirmatory study. Lancet 2017;389:815-22. [Crossref] [PubMed]
- Adhyam M, Gupta AK. A Review on the Clinical Utility of PSA in Cancer Prostate. Indian J Surg Oncol 2012;3:120-9. [Crossref] [PubMed]
- Lee HH, Kim SH. Review of non-invasive urinary biomarkers in bladder cancer. Transl Cancer Res 2020;9:6554-64. [Crossref] [PubMed]
- Cheng THT, Jiang P, Teoh JYC, et al. Noninvasive Detection of Bladder Cancer by Shallow-Depth Genome-Wide Bisulfite Sequencing of Urinary Cell-Free DNA for Methylation and Copy Number Profiling. Clin Chem 2019;65:927-36. [Crossref] [PubMed]
Cite this article as: Chehade L, Abdel Sater AH, Kourie HR, Mukherji D, Shamseddine A. Clinical implications of liquid biopsies in urogenital tumors: a narrative review. Precis Cancer Med 2022;5:24.