
Next-generation sequencing using liquid biopsy in the care of patients with ALK-rearranged non-small cell lung cancer: a focus on lorlatinib
Introduction
The treatment of patients with anaplastic lymphoma kinase (ALK) rearranged lung cancer has improved in the last years mainly due to the understanding of the acquired mechanisms of resistance to early generation ALK tyrosine kinase inhibitors (TKI) prompting the development of new generation ALK inhibitors.
Several mechanisms of resistance to ALK inhibitors have been characterized like ALK resistance mutations and off-target bypass mechanisms of resistance. However, unlike T790M mutation status to select treatment with osimertinib in EGFR-mutant lung cancer, the selection of new generation ALK inhibitors after crizotinib did not include mandatory biomarker assessment of resistance mechanisms to guide treatment to subsequent ALK inhibitors (1,2). In patients with metastatic lung cancer, longitudinal tissue biopsies are difficult to perform and can potentially lead to clinical complications. Moreover, the biopsy of a specific progressing tumor lesion may not be representative of all the biological mechanisms that drive resistance to ALK inhibitors, which can be heterogeneous, spatially and temporarily (3,4).
The rapid development and clinical implementation of circulating tumor DNA (ctDNA) next-generation sequencing (NGS), allows to longitudinally interrogate the patients tumor biology, asses genomic tumor heterogeneity, and potentially identify resistance mechanisms that can guide treatment decisions (5). Implementing treatment decisions in patients with lung cancer using cfDNA was first done with the detection of the epidermal growth factor receptor (EGFR) T790M gatekeeper mutation using standard real-time PCR or digital droplet PCR assays (6). This has changed the diagnostic algorithm to guide the selection of following treatments in patients experiencing progression on first- and second-generation EGFR inhibitors, starting with a liquid biopsy for EGFR T790M detection, and if negative pursuing a tissue biopsy to rule out potential false-negative results (7). The EGFR T790M accounts for about 40–50% of resistance to first- and second-generation EGFR inhibitors (8). Resistance mutations to third-generation EGFR inhibitors, such as C797S have also been described using liquid biopsy (9). Moreover, characterization of C797S mutation in cis or trans with T790M, has treatment implications, with reports of response to the combination of first and third generation EGFR inhibitors in patients with mutations in trans (10). This is different in the setting of ALK TKI resistance in which multiple different ALK kinase domain resistant mutations can be acquired, for which NGS is required to map exons 20 to 28 that codify for this domain (11).
Lorlatinib is a third-generation ALK TKI recently approved for the treatment of patients that experience disease progression after a first- and second-generation ALK inhibitors or second generation ALK TKIs upfront (12).Lorlatinib was designed to overcome resistance by all known single and acquired ALK resistance mutations, including the solvent front G1202R mutation that mediates resistance to all first- and second-generation ALK TKIs (13). Unfortunately, even with the development of this highly potent ALK inhibitor, all patients will eventually experience disease progression due to the adaptation of cancer cells to lorlatinib selective pressure. To date, there are no precise biomarkers to adequately predict which patients will benefit the most from treatment with lorlatinib, and there is scarce data on lorlatinib resistance mechanisms and ways to prevent it.
In this review we analyze the current and potential role of liquid biopsy NGS as a biomarker for treatment selection after progression with second-generation ALK inhibitors and as a novel strategy to study lorlatinib resistance mechanism.
Overview on ALK rearrangements and ALK inhibitors in lung cancer
ALK rearrangements occur in about 3–6% of advanced lung adenocarcinomas (14,15). The fusion protein contains the complete ALK kinase domain, and the fusion partner mediates homodimerization of the fused protein to induce ALK transactivation, phosphorylation, and recruitment of adaptor proteins that trigger downstream oncogenic signaling (16,17).
Echinoderm microtubule-associated protein-like 4 (EML4) gene is the most common ALK fusion partner, present in 81% of ALK-positive NSCLC (18-20). However, multiple other gene partners have been described including KIF5B, STRN, SLC2A, amongst others (17). The EML4 breaking point in EML4-ALK rearrangements defines different fusion variants, of which variant 1 is the most common, accounting for 43% of cases, followed by variant 3 in about 40% (21,22). Shorter variants that do not contain the EML4 TAPE domain, like variant 3 and 5, are more stable proteins. There is controversy regarding the prognostic role of different EML4-ALK variants in patients treated with crizotinib and second-generation ALK inhibitors (22-26).
There are currently several ALK inhibitors that have been granted regulatory approval, the first-generation ALK inhibitor crizotinib, the second-generation ALK TKIs ceritinib, alectinib, and brigatinib, and the third-generation inhibitor lorlatinib. Currently, there are two different approved treatment strategies: first-line treatment with crizotinib followed by second-generation ALK inhibitors or frontline treatment with a second-generation ALK inhibitor, followed in both strategies by lorlatinib at the time of disease progression (10,23-26). In both scenarios, starting treatment with a first- or second-generation ALK inhibitor can result in a 4-year overall survival (OS) rate of about 50%, proving that patients with ALK-rearranged lung cancer treated with ALK TKIs can most likely have prolonged survival when treated with sequential lines of ALK inhibition (27,28). The median progression-free survival (PFS) for patients treated with upfront crizotinib is about 10.9 months, and the median PFS with second-generation inhibitors given sequentially ranges from 5.4 to 15.6 months (27-29). First-line treatment with second-generation ALK inhibitors like alectinib and brigatinib, confer prolonged progression-free survival and intracranial disease control compared to crizotinib (29,30). The median PFS reported in the ALEX study, which compared first-line treatment with alectinib to crizotinib in patients with metastatic ALK-rearranged NSCLC was 34.8 months compared to 10.9 months (HR: 0.43 95% CI: 0.32–0.58) (26). In the ALTA1L study, comparing brigatinib to crizotinib in the frontline setting, the median PFS was also significantly superior with the second-generation ALK inhibitor (24.0 vs. 11.0 months; HR 0.49, P=0.001) (29). Both alectinib and brigatinib are currently standard first-line treatment options based on these clinical trials.
New generation ALK inhibitors have been designed to overcome resistance to crizotinib, mainly “on-target” resistance due to the acquisition of secondary ALK kinase domain mutations that impede crizotinib inhibition of the kinase domain by modifying the kinase structure (e.g., ALK L1196M gatekeeper mutation) or by enhancing the kinase ATP affinity (e.g., ALK F1174L) (31,32). ALK kinase domain mutations involved in crizotinib resistance include L1152P, C1156Y, I1171T, F1174C/L/V, L1196M, G1202R, D1203N, S1206C/Y, E1210K, and G1269A (1,32-36). Of these mutations, the most common are the gatekeeper L1196M and the G1269A ATP-pocket mutation, and cancer cells that harbor ALK-rearrangements with these mutations are highly susceptible to all second-generation ALK inhibitors (1,37). Contrarily, the solvent front ALK G1202R mutation, present in about 2% of crizotinib samples, is the most common ALK-dependent resistance mechanism in patients treated with second-generation ALK inhibitors (~40%) (1,38). However, the spectrum of activity against other crizotinib resistant mutations differs between ceritinib, alectinib and brigatinib. Ceritinib is inactive in the setting of ALK I1151X, L1152P, C1156Y and F1174X mutations (1,31). Alectinib is active against these mutations but does not inhibit ALK in the context of I1171X, and V1180L mutations (39,40). Brigatinib, however, is active against all non-ALK G1202R mutations including those for which alectinib and ceritinib lack inhibitory activity (41).
The type of EML4-ALK variant has been associated with different patterns of ALK resistance mutation acquisition. In a multicenter analysis using tissue NGS to identify EML4-ALK variants and resistance mechanism, ALK mutations at progression with second-generation ALK TKIs were more frequent in variant 3 fusions (66%) compared to variant 1 fusions (42%), though this difference was not statistically significant (22). However, the acquisition of ALK G1202R mutations at resistance was significantly enriched in variant 3 rearrangements compared to variant 1 (44% vs. 0%, P=0.001). This was further validated in a larger data set from Foundation Medicine, showing that ALK resistance mutations were significantly more frequent in variant 3 compared to variant 1 EML4-ALK fusions, including the ALK G1202R mutation (32% vs. 0%, P=0.001) (22).
In addition to ALK kinase domain mutations, ALK amplification causes resistance to crizotinib but can be overcome with more potent second-generation inhibitors (34). ALK amplification has not been reported as a resistance mechanism to ceritinib, alectinib nor brigatinib.
Lorlatinib: the third generation ALK inhibitor
Lorlatinib is a potent third-generation ATP competitive ALK inhibitor and also active against ROS1-rearranged lung cancers. Its pharmacological development included the design of a macrocyclic molecule based on the crizotinib structure, modified to specifically bind to ALK in the presence of all known single ALK resistance mutations (13). In preclinical studies using in vitro kinase assays, lorlatinib showed higher ALK inhibitory potencies than crizotinib, ceritinib, and alectinib (13). Besides, in Ba/F3 cells expressing EML4-ALK with the G1202R mutation, and in patient-derived cell lines that harbored the ALK G1202R mutation, lorlatinib induced cell death in vitro (IC50 value ~63 nM). Also, lorlatinib, like alectinib and brigatinib, is not a substrate of the p-glycoprotein efflux system, leading to high levels of central nervous system penetration and concentration (13). In brain orthotopic mice models the free fraction of lorlatinib in the central nervous systems compared to plasma was 4-fold higher than crizotinib. Preclinical studies using ceritinib resistant patient-derived cell lines showed that lorlatinib was selectively active in models in which ALK resistance mutations were present and not in models that did not harbor ALK mutations in which bypass alterations or other off-target resistance mechanisms could be present (1). Thus, in vitro studies initially suggested that lorlatinib was solely active against tumors that harbored ALK-dependent resistance mechanisms.
The potency of ALK inhibition and the high brain barrier penetration was confirmed in the phase I study of lorlatinib including 41 pretreated patients of which 72% had brain metastasis. In the pharmacokinetic analysis, the mean cerebral-spinal fluid concentration of lorlatinib was 75% of the plasma concentration (42). The pivotal results of this phase I study showed that about 46% of patients experienced an overall respone, including 57% of patients that received one previous ALK inhibitor and an objective response rate of 42% in patients previously treated with two or more lines of ALK TKIs (42). In patients with measurable and non-measurable brain metastasis, the intracranial response rate was 31%.
Early biomarker assessments in the phase I trial concurred with the preclinical in vitro data supporting the role of lorlatinib in tumors harboring ALK resistant mutations. All nine patients with detectable ALK resistance mutations in tumor samples experienced tumor regression with lorlatinib, including five tumors with detectable ALK G1202R and G1202del mutations. However, in three patients in which ALK mutations were not detected, there was no evidence of clinical response.
The clinical development continued with the phase II multicohort expansion study including treatment naïve patients (EXP1), patients previously treated with crizotinib only (EXP2), crizotinib and chemotherapy (EXP3A), second-generation ALK inhibitor +/− chemotherapy (EXP3B), two lines of ALK TKIs +/− chemotherapy (EXP4), 3 prior lines of ALK TKIs +/− chemotherapy (EXP5) (12). This trial design allowed to adequately explore different clinical scenarios in which lorlatinib could have a role. Lorlatinib given as a first-line therapy in 30 patients (EXP1) conveyed an objective response rate (ORR) of 90% with a median PFS that was not reached (NR) (95% CI: 11.4 to NR) and intracranial responses (ICR) in 66.7% of patients. In 59 patients previously treated with crizotinib +/− chemotherapy (EXP2-3A) the ORR was 69.5%, median PFS was not reached (95% CI: 12.5 to NR) and the ICR rate was 87%. In patients previously treated with one second-generation ALK inhibitor (EXP3B) and in patients treated with two or more ALK TKIs (EXP4-5) the ORR was 32.1% and 38.7%, respectively. The median PFS was 5.5 months (95% CI: 2.7–9.0) and 6.9 months (95% CI: 5.4–9.5) and ICR rates were 55.6% and 53.1%, respectively. This study led to the FDA approval of lorlatinib in the setting of disease progression on a second-generation ALK inhibitor, given as a first-line treatment or after progression on crizotinib and a second-generation ALK inhibitor in the second line.
Real-world data also supports the efficacy of lorlatinib in patients previously treated with first- and/or second-generation ALK inhibitors that received lorlatinib through expanded access programs (43). Among individuals treated with two previous ALK TKIs the objective response rate was 42% and the median PFS was not reached (95% CI: 4.5 to NR), in patients treated with more than two ALK TKIs the ORR was 35% and the median PFS was 11.2 months. Intracranial responses were observed in 52% of patients overall, also contributing to the external validation of the phase I/II trial of lorlatinib.
The phase II study also provided some early evidence of the potential activity of lorlatinib in the first-line setting, also supported by clinical evidence of enhanced activity with first-line alectinib in the ALEX study and brigatinib in the ALTA1L trial (29,30). This led to the design of the phase III CROWN study, comparing front line therapy with lorlatinib to crizotinib in 296 patients with ALK-rearranged lung cancer (NCT03052608). The first reported results of this study show that upfront treatment with lorlatinib significantly prolongs PFS compared to crizotinib [median PFS: not reached vs. 9.3 months; HR: 0.28 (95% CI: 0.19–0.41)]. Moreover, objective response was significantly higher in the lorlatinib group (79% vs. 58%), with 70% of patients maintaining responses at 12 months. In addition, lorlatinib treatment resulted in higher intracranial responses (66% vs. 20%) and central nervous system (CNS) time to progression, 96% of patients without CNS progression at 12 months with lorlatinib vs. 60% with crizotinib (44). This subsequently led to the FDA-approval of lorlatinib as a first-line treatment option in treatment naïve patients.
Preclinical and clinical evidence on lorlatinib resistance mechanisms
There is an increasing amount of evidence on lorlatinib resistance based on preclinical studies and translational research from patients’ samples during early phase lorlatinib development. The first report of resistance to lorlatinib was done by extensively studying the evolution of on-target mechanisms in a patient whose tumor acquired sequential ALK mutations during exposure to crizotinib, ceritinib, and lorlatinib (45). After first-line treatment with crizotinib an ALK C1156Y mutation was detected on a lymph node biopsy at disease progression. As previously mentioned, this mutation confers resistance to crizotinib and ceritinib. The patient continued treatment with ceritinib experiencing primary progression. The patient was later included in the phase I study of lorlatinib, achieving a partial response that lasted for 8 months. Eventually, the patient experienced disease progression in the liver and a new liver biopsy showed the presence of both the previously identified ALK C1156Y mutation and an additional ALK L1198F mutation. Both mutations were present at similar allele frequencies and were confirmed to be in the same EML4-ALK allele by subcloning PCR products into pCR4-TOPO vectors and performing bacterial colony sequencing. Clonal evolution analysis using whole exome sequencing revealed that clones harboring both mutations arose from ALK C1156Y mutant cells.
The sequential acquisition of two or more mutations in the ALK kinase domain is now called “compound mutations”. In vitro modeling using Ba/F3 cells harboring single and compound mutations in EML4-ALK infected cells showed that the presence of the ALK C1156Y/L1198F compound mutation resulted in lorlatinib resistance, impeding drug binding to the mutant kinase domain. Interestingly, the presence of phenylalanine in codon 1198 modified the structural conformation of the kinase to favor crizotinib binding, counterbalancing the increased kinase ATP affinity induced by the ALK C1156Y mutation that would normally cause crizotinib resistance. The patient was treated later with crizotinib experiencing a partial response lasting for almost six months. This first study, by the in-depth characterization of the patient’s tumor biological evolution, resulted in the identification of compound mutations as a novel mechanism of resistance to lorlatinib, and at the same time, showed that specific compound mutations can potentially resensitize cancer cells to previous generations of ALK inhibitors.
Compound mutations have also been characterized in brigatinib resistant tumors, like the ALK D1203N/E1210K compound mutation, however, in vitro models carrying this compound mutation retain sensitivity to lorlatinib (1).This further portrays the need to fully characterize compound mutations to include a new repertoire of on-target alterations that can potentially aid in the selection of active ALK inhibitors in this setting.
To predict which on-target single or compound mutations could confer resistance to lorlatinib, N-ethyl-N-nitrosourea (ENU) mutagenesis screens using Ba/F3 cells that harbor the EML4-ALK fusion were performed in two studies. When exposing Ba/F3 cells to ENU and treatment with lorlatinib alone, there were no resistant clones emerging, showing that upfront treatment with lorlatinib could suppress the emergence of single mutant resistant cells in vitro (46). Differently, in Ba/F3 cells containing common resistance mutations to first- or second-generation ALK inhibitors, exposed to ENU and lorlatinib, multiple different compound mutations emerged, validating that the acquisition of more than one ALK mutations in cis is required to convey resistance to lorlatinib (46,47).
Compound mutations have been detected in about 35% of patients at the time of progression with lorlatinib (46). Several compound mutations have been identified in patients to confer resistance to lorlatinib like I1171N/L1198F, G1202R/G1269A, G1202R/L1196M; G1202R/F1174L (46-48) (Figure 1A, Table 1). Interestingly, also compound mutations acquired with first and second-generation ALK inhibitors can also confer primary resistance to lorlatinib, like the L1196M/D1203N compound mutation, reported in a patient whose tumor acquired the L1196M mutation on crizotinib and sequentially the D1203N mutation on ceritinib (48). In addition, like in the case of the C1156Y/L1198F mutation, the compound I1171N/L1256F mutation was found in vitro by ENU mutagenesis to cause lorlatinib resistance but resensitized these cells to alectinib, even when the I1171N mutation alone confers high levels of resistance to alectinib (47). Moreover, the ALK l1256F mutation alone can cause lorlatinib resistance and, so far, is the sole single ALK mutation reported to cause lorlatinib resistance in vitro. The ALK L1256F mutation is analogous to the ROS1 L2086F which has been reported as a resistance mechanism in a patient with ROS1-rearranged lung cancer that experienced disease progression with lorlatinib (50).
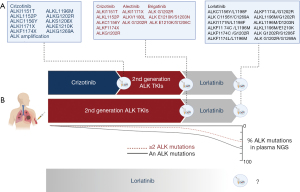
Table 1
| Compound mutation | Lorlatinib | Alectinib | Crizotinib | Citations |
|---|---|---|---|---|
| L1198F/C1156Y | R | R | S | (45) |
| L1198F/L1196M | R | R | S | (46) |
| L1198F/G1202R | R | R | S | (47) |
| I1171N/L1198F | R | R | S | (47) |
| I1171N/L1256F | R | S | R | (47) |
| I1171N/L1196M | R | R | R | (47) |
| I1171N/G1296A | R | R | R | (47) |
| I1171S/G1269A | R | R | R | (49) |
| C1156Y/C1269A | S | R | R | (48) |
| L1196M/D1203N | R | R | R | (48) |
| L1196M/G1202R | R | R | R | (46,47) |
| F1174C/G1202R | R | R | R | (48) |
| F1174L/G1202R | R | R | R | (47) |
| G1202R/G1269A | R | R | R | (46) |
R, resistant; S, sensitive.
Not all compound mutations that can be found on tissue or plasma samples at the time of progression to lorlatinib are the cause of lorlatinib resistance. In a patient previously treated with crizotinib, an ALK C1156Y/G1269A compound mutation was detected using tissue NGS in a tumor biopsy at the time of lorlatinib resistance, however, this compound mutation did not cause lorlatinib resistance by in vitro characterization. This suggests that other resistance mechanisms can drive tumor growth even in the presence of compound mutations (48). Given that some ALK compound mutations can be targeted with earlier generation ALK inhibitors as previously shown, and that not all compound mutations will cause lorlatinib resistance, there is a need to fully characterize the spectrum of compound mutations to improve treatment strategies for patients in the future.
ALK-independent or “off-target” resistance mechanisms have also been reported to mediate lorlatinib resistance. MET amplification has been recently identified in about 15% of tumor biopsies from patients progressing on second-generation ALK inhibitors (12%) and lorlatinib (22%) (51). MET amplification was commonly found in tumors from patients treated upfront with second-generation ALK inhibitors compared to patients receiving first-line treatment with crizotinib, which is also a type Ia MET inhibitor. In a few cases, combining ALK-MET inhibition led to clinical responses in patients (51). Acquired MET amplification has been well known to cause resistance to EGFR inhibitors, and clinical trials combining EGFR TKIs and selective MET inhibitors, like osimertinib and savolitinib in the TATTON trial, have shown encouraging clinical results (52). Clinical trials aiming to overcome MET-driven resistance in patients with ALK-rearranged NSCLC are highly needed. Other ALK independent resistance mechanisms described include NF2 loss of function mutations, SRC activation and epithelial-mesenchymal transition in vitro (48). Histologic transformation can also occur, and neuroendocrine transformation has been reported to confer resistance to lorlatinib in a patient (53).
The role of liquid biopsy NGS to study response and resistance to lorlatinib
Plasma circulating tumor DNA (ctDNA) NGS has become a more widely available molecular biology technique to interrogate cancer genomics through a blood draw without requiring tissue biopsy (7). Liquid biopsies can be informative in several scenarios in the setting of ALK-rearranged NSCLC: at diagnosis in treatment naïve patients, to monitor response and progression during treatment with targeted therapies, to select sequential treatments according to resistance mechanisms in previous lines of therapy, and finally, at the time of disease progression to study novel resistance mechanisms (Figure 1A). Few studies have focused on the role of liquid biopsies to predict lorlatinib activity and to depict resistance mechanisms.
Liquid biopsy is an alternative tool to study ALK fusions at diagnosis when tissue is unavailable, though the sensitivity of NGS in plasma to detect ALK fusions ranges from 67% to 91% (54,55). Patients in which ALK rearrangements are detected by liquid biopsies, as expected, also benefit from treatment with ALK inhibitors. In the BFAST trial in 2,219 patients screened using foundation liquid NGS assay, ALK-rearrangements were found in 5.4% of plasma samples. Patients with ALK-rearranged NSCLC detected by liquid biopsies achieved an ORR of 92% and a 12-month PFS rate of 78.4%.
Pretreatment determination of the type of EML4-ALK rearrangements might have clinical implications in the future. As previously addressed, plasma biomarker study of the ALEX trial showed that in patients with EML4-ALK rearrangement detected in plasma the median PFS with alectinib was 34.8 months for variant 1, 24.8 months for variant 2, and 17.7 months in variant 3, though this difference was not statistically significant (26). However, in a biomarker analysis of the phase III ALTA1L study comparing frontline treatment with brigatinib to crizotinib, PFS was significantly shorter in patients with variant 3 EML4-ALK rearrangements compared to variant 1 treated with brigatinib [HR 2.38 (95% CI: 1.04–5.5)] and crizotinib [HR 2.96 (95% CI: 1.44–6.09). This could be explained by the fact that EML4-ALK variant 3 tumors have higher rates of acquired ALK resistance mutations (44.4% variant 1 vs. 75% variant 3) and ALK G1202R mutations (0% in variant 1(0/9) vs. 50% (4/8) in variant 3) compared to variant 1 EML4-ALK fusions (22). In another study evaluating the use of plasma NGS with InVisionFirst-Lung assay from Inivata, 37% of EML4-ALK variant 3 fusions had ALK kinase domain mutations compared to 13% with variant 2and 0% in variant 1 fusions, and all G1202R mutations were seen in variant 3 EML4-ALK rearrangements (54).
The role of liquid biopsies has also been studied aiming to predict clinical benefit with lorlatinib in patients pre-treated with first- and/or second-generation ALK TKIs (Table 2). In the lorlatinib registrational phase II trial, plasma samples were obtained before treatment with lorlatinib and analyzed using Guardant360 NGS (38). Tissue biopsies were also conducted and analyzed using the Ion Torrent PGM platform. Among 198 patients enrolled in the trial that received prior ALK directed therapies (EXP2-5), 59 received only prior therapy with crizotinib, and 139 patients,prior therapy with one or more second-generation ALK TKI. Across all 189 patients with available plasma, 21% of samples had no detectable cell-free DNA (cfDNA) and in 24% of samples, one or more ALK kinase domain mutations were found. From tissue biopsies, NGS was done on 78%, so 22% were inadequate for NGS analysis, a similar rate of sequencing failure compared to plasma NGS. In adequate tissue samples, ALK mutations were found in 47% of cases. Among patients that received prior second-generation ALK inhibitors, the ALK G1202R mutation was found in 53% of cfDNA plasma samples and in 55% of tissue samples. Using tissue biopsies from lorlatinib pretreatment samples as a reference, the sensitivity of plasma NGS for ALK mutations was 61% and the specificity was 82%, with an overall accuracy for plasma NGS of 73%, which needs further improvement.
Table 2
| Study | Number of patients | ORR | mPFS (months) | NGS platform | |||
|---|---|---|---|---|---|---|---|
| Post crizotinib | Post 2nd generation | Post crizotinib | Post 2nd generation | ||||
| Shaw et al. (38) 2019 | Tissue NGS adequate for NGS: | Ion torrent PGM platform | |||||
| ALK mutations: 40 (24%) | 73% | 69% | NR (95% CI: 2.6 to NR) | 11 (95% CI: 6.9 to NR) | |||
| No ALK mutations: 124 (76%) | 74% | 32% | 12.5 (95% CI: 0.4–3.9) | 5.4 (95% CI: 3.9–6.9) | |||
| Tissue Inadequate for NGS: 27 | HR: 1.38(95% CI: 0.48–3.98) | HR: 0.47 (95% CI: 0.27–0.83) | |||||
| Plasma NGS: | Guardant 360 | ||||||
| ALK mutations: 45 (24%) | 73% | 62% | NR (95% CI: 1.7–NR) | 7.3 (95% CI: 4.1–13.1) | |||
| No ALK mutations: 104 (55%) | 75% | 32% | 12.5 (95% CI: 6.9–NR) | 5.4 (95% CI: 3.9–6.9) | |||
| No detectable cfDNA: 40 (21%) | HR: 1.03 (95% CI: 0.39–2.69) | HR: 0.81 (95% CI: 0.50–1.31) | |||||
NGS, next-generation sequencing; ORR, objective response rate; mPFS, median progression-free survival.
Most importantly, given the preclinical data showing that lorlatinib was most effective in patients with on-target resistance, this biomarker driven study compared the outcomes of patients with detectable ALK mutations using plasma and tissue NGS to patients without detectable alterations. The objective response rate was higher in patients with ALK mutations detected by plasma (62% vs. 32%) and “de novo” tissue NGS (69% vs. 31%) compared to patients without detectable ALK mutations (38). However, there were no significant differences in PFS between patients with and without detectable ALK mutations in plasma, median PFS 7.3 vs. 5.5 months [HR: 0.81 (95% CI: 0.5–1.31)]. Among patients with tissue NGS, median PFS was significantly prolonged among patients with detectable ALK mutations, especially in “de novo samples” with median PFS 11.0 months compared to 4.0 months in patients without detectable ALK mutations in tissue biopsies [HR:0.20 (95% CI: 0.10–0.40)]. The difference in outcomes between plasma and tissue NGS could be explained by the lower sensitivity of plasma NGS to identify ALK resistance mutations before lorlatinib treatment, so several patients with ALK mutations detectable in tissue NGS but not in plasma may enrich the outcomes of the group of patients without detectable ALK mutations by plasma due to the false-negative rate of this technique. According to these results, though detecting ALK mutations by plasma or tissue NGS is related to higher response rates, about 30% of patients without detectable plasma mutations will respond to treatment with lorlatinib, thus for the moment, there is not a role for plasma or tissue genotyping as a selection biomarker in this setting. However, plasma or tissue NGS can be informative on the likelihood of response in a patient according to the ALK mutation status and provide information on resistance mechanisms. Plasma NGS is highly convenient to avoid new tissue biopsies, but the lack of predictive role limits its mandatory use in the clinical practice.
Another study comparing tissue and plasma NGS using Guardant 360 in patients with ALK-rearranged NSCLC showed that paired tissue and plasma samples had a similar rate of ALK mutation detection, at the time of disease progression on alectinib of 63% and 67%, respectively (56). However, plasma NGS was more likely to detect multiple ALK kinase domain mutations in this setting (24% vs. 2%, P=0.004), proving that plasma NGS may be more informative of polyclonal on-target resistance or the acquisition of a compound mutation after first- and second-generation ALK TKI.
Compound mutations are infrequently found after first- and second generation ALK inhibitors, but have been reported to cause primary resistance to lorlatinib. This is the case of a patient whose tumor acquired the gatekeeper L1196M mutation with crizotinib and received second line treatment with ceritinib, at the time of disease progression, only the L1196M mutation was found in tissue NGS, however plasma NGS using the Inivata InVisionFirst-Lung assay, detected both the L1196M and a solvent front D1203N mutation, and due to proximity of these mutations they were found to be in cis (48). The patient experienced primary progression with lorlatinib, due to the effect of this compound mutation in halting lorlatinib binding to the kinase domain, conferring a 300-fold shift in the IC50 of Ba/F3 cells harboring these compound mutations treated with lorlatinib compared to single mutant cells. In this case, plasma NGS was more informative detecting this compound mutation by capturing tumor heterogeneity, which was not observed with tissue NGS of a single site biopsy. However, the development of a compound mutations is most likely a rare event during resistance to first- and second-generation ALK inhibitors.
As we mentioned earlier, acquired compound mutations are a major determinant of ALK-dependent lorlatinib resistance. Nonetheless, confirming that both mutations are in cis solely by targeted plasma or tissue NGS is complicated, mainly dependent on the proximity and inclusion of both mutations in the same NGS read. ALK resistance mutations are more commonly detected after progression to lorlatinib using plasma NGS (Figure 1B). In a study by Mezquita and colleagues, 43% of plasma samples undergoing plasma NGS in patients experiencing disease progression on lorlatinib had detectable ALK mutations compared to 29% of patients progressing on second-generation ALK TKIs and 11% with crizotinib (54)(Table 3). In this study, “complex” ALK mutations referred to the detection of more than one ALK mutation including compound mutations and multiple mutation in which determining the allelic distribution was not possible. Among three patients with paired tissue and plasma samples in which complex ALK mutations were detected by this later method, there was discordance in the type of mutations detected and in the number of mutations, which was higher for plasma genotyping. In one case, in which an ALK G1202R and F1174L mutations were detected using tissue NGS and confirmed to be in cis by TOPO-TA cloning of DNA fragments of the kinase domain, multiple other mutations were additionally found using plasma NGS, including: C1156Y, T1151M, G1269A and S1206F. Of all these mutations emerging at lorlatinib resistance, solely the G1202R/F1174L and by proximity, the G1202R/S1206F mutations were confirmed to be in cis, reflecting also that compound mutations can be acquired in different tumor cell clones, and become a polyclonal event, difficult to target (48,54). Reflecting the complexity of on-target resistance with lorlatinib, in the same patient in addition to the multiple ALK mutations detected, NGS of circulating tumor cells found a G1202R/F1174C mutation that was not detected on tissue nor plasma NGS (57).
Table 3
| Study | Number of patients | ALK mutations | Complex mutations (≥2 ALK mutations) | Compound ALK mutations | NGS platform |
|---|---|---|---|---|---|
| Dagogo-Jack et al. (56) 2019 | Tissue NGS: | Not reported | SNaPshot NGS | ||
| Post 2nd generation TKI: 41 | 26 (63%) | 1 (2%) | Foundation One | ||
| Post lorlatinib: 32 | 12 (38%) | 9 (28%) | DFCI Oncopanel | ||
| MSK Impact | |||||
| Plasma NGS: | Guardant 360 | ||||
| Post 2nd generation TKI: 70 | 46 (66%) | 16 (23%) | 5/5 (100%) | ||
| Post lorlatinib: 29 | 22 (76%) | 14 (48%) | 3/6 (50%) | ||
| Mezquita et al. (54) 2020 | Plasma NGS: | Not reported | InVisionFirst-Lung | ||
| Post crizotinib: 36 | 4 (11%) | 3 (8%) | |||
| Post 2nd generation TKI: 31 | 9 (31%) | 1 (3%) | |||
| Post lorlatinib: 7 | 3 (43%) | 3 (43%) |
FISH, fluorescence in situ hybridization; CEP7, centromere chromosome 7; NGS, next-generation sequencing.
In the largest study of resistance to lorlatinib using plasma NGS, Dagogo-Jack and colleagues studied the role of plasma cfDNA genotyping at the time of progression on second-generation ALK TKIs and lorlatinib using the Guardant360 assay (56) (Table 3). ALK resistance mutations were seen in 66% (46/70) of patients after second-generation ALK inhibitors, and in 76% (22/29) of patients progressing on lorlatinib. The detection rate of ≥2 concomitant ALK mutations was doubled at progression on lorlatinib compared to second-generation ALK inhibitors, 48% compared to 23%, respectively (P=0.017). In patients that received lorlatinib after a second-generation ALK inhibitor, 53% (8/15) acquired a new ALK resistance mutation during the course of therapy. Moreover, in patients with paired tissue and plasma genotyping at the time of resistance to lorlatinib, liquid biopsy NGS was more likely to detect ALK mutations compared to tissue NGS, 76% versus 38%, respectively (P=0.004). Also, plasma NGS detected ≥2 ALK mutations at higher rates in plasma compared to tissue (48% versus 28%) though this was not statistically significant.
In this study, among five samples obtained at lorlatinib progression with ≥2 ALK mutations that were close enough to asses allelic distribution, all were identified as compound mutations, including one patient with 3 different ALK compound mutations (G1202R/L1196M, L1196M/F1174L and L1196M/F1174C) again supporting the new concept that distinct compound mutations can be present in different clones emerging under selective pressure with lorlatinib (56). Among patients with newly acquired ALK resistance mutations, the ALK D1203N solvent front mutation was more commonly acquired with lorlatinib than with second-generation ALK inhibitors.
Characterization of lorlatinib resistance by liquid biopsy NGS may have clinical implications in the near future with the development of new generations of ALK inhibitors that can bind and block ALK phosphorylation in the context of specific compound mutations. TPX-0131 (turning point therapeutics) is a novel macrocyclic ALK inhibitor that can bind to the ATP binding pocket in the presence of a range of compound mutation combinations that contain the ALK G1202R mutation, and inhibit ALK in vitro and in xenograft models (58). Some of these compound mutations include ALK G1202R/C1156Y, G1202R/L1196M, G1202R/C1198F, G1202R/G1269A. However, this drug does not inhibit ALK in the presence of I1171X mutations that are a common resistance mechanism to alectinib. If the clinical development of this drug is pursued, predictive biomarkers of response will be required to appropriately select patients.
Off-target lorlatinib resistance can also be assessed using plasma NGS. MET amplification was detected in 13% of patients assessed by FISH or NGS in tissue samples, including 22% of post-lorlatnib biopsies (5/23) (51) (Table 4). Among 106 plasma samples, MET focal amplification (defined in this study as absolute MET copies ≥2.1 based on the validation study of plasma comprehensive cancer genotyping assay (59) was detected in seven cases (6.6%), including 17% of plasma samples obtained at lorlatinib resistance (5/29). Among 23 patients with paired tissue and plasma genotyping, the sensitivity, specificity, and positive predictive value of plasma NGS using Guardant360 to detect MET amplification was 100%, 95%, and 80%, respectively. Other off-target resistance mechanisms like KRAS amplification and a PI3KCA E545K mutation were also present concomitantly with MET amplification in a patient, showing that several off-target resistance mechanisms can also coexist (51). Other putative bypass track resistance mechanisms have also been described at resistance to ALK inhibitors including lorlatinib, like KRAS mutations and PTEN mutations. However, the impact of off-target mutations detected prior to lorlatinib treatment has not been reported so far, and larger studies are required to fully depict the range of ALK-independent lorlatinib resistance.
Table 4
| Study | Definition of MET amplification | MET amplification | Co-occurring ALK mutations | NGS platform |
|---|---|---|---|---|
| Dagogo-Jack et al. (51) 2020 | Tissue: | Post: | ALK I1171N | Foundation one |
| FISH: MET/CEP7 ≥2.2 | 2nd generation: 6/52 (12%) | |||
| Foundation one: MET copy number ≥6 | Lorlatinib 5/23 (22%) | |||
| Plasma: | Post: | ALK I1171N | Guardant 360 | |
| Guardant 360: MET copy number ≥2.1 | 2nd generation: 2/77 (3%) | ALK L1196M | ||
| Lorlatinib: 5/29 (17%) | ALK L1196M | |||
| ALK L1196M |
NGS, next-generation sequencing.
Conclusions
Plasma cell free DNA NGS is becoming a widely adopted molecular biology technique in the diagnosis and treatment of patients with lung cancer. Lorlatinib is the most recent approved ALK inhibitor, capable of potentially overcoming resistance to first- and second-generation ALK inhibitors, constituting the last available line of ALK directed therapies so far. For the moment, liquid biopsy NGS has limitations to properly select patients prior to lorlatinib initiation but, per contrary, seems to convey more information at the time of disease progression, being highly informative on lorlatinib resistance mechanisms. In the future, liquid biopsies could potentially be useful to guide upfront treatment selection and subsequent therapies according to the type of resistance mechanisms detected at the time of disease progression, in light of the development of novel third-generation ALK inhibitors.
Acknowledgments
Figure 1: created with BioRender.com.
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Jesús Corral, Laura Mezquita and Ernest Nadal) for the series “ALK and ROS-1 NSCLC Patients Treatment Approach Based on Genomic Profile by Liquid Biopsy” published in Precision Cancer Medicine. The article has undergone external peer review.
Peer Review File: Available at https://dx.doi.org/10.21037/pcm-20-59
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/pcm-20-59. The series “ALK and ROS-1 NSCLC Patients Treatment Approach Based on Genomic Profile by Liquid Biopsy” was commissioned by the editorial office without any funding or sponsorship. Dr. AR serves as an unpaid editorial board member of Precision Cancer Medicine from Aug 2020 to Jul 2022. Dr. GR reports grants from AMGEN, personal fees from ROCHE, personal fees from PFIZER, Bayer, BMS, MSD, Pfizer, Takeda, and personal fees from AMGEN, from null, during the conduct of the study. AFC disclose financial research support from Merck Sharp & Dohme, Boehringer Ingelheim, Roche, Bristol-Myers Squibb and The Foundation for Clinical and Applied Cancer Research - FICMAC. Additionally, he was linked and received honoraria as advisor, participate in speakers’ bureau and gave expert testimony to Merck Sharp & Dohme, Boehringer Ingelheim, Roche, Bristol-Myers Squibb, Pfizer, Novartis, Celldex Therapeutics, Foundation Medicine, Eli Lilly and Foundation for Clinical and Applied Cancer Research - FICMAC. Dr. AR reports consultancy/advisory board role for Astra Zeneca, MSD, and Novartis. Dr. CR reports Consulting fees from: ARCHER, Inivata, BMS, Novartis, Boston Pharmaceuticals, EMD Serono, Astra Zeneca, Pfizer, Mirati, Eisai. Grants from Pfizer and fees for non-CME/CE services from Astra Zeneca, Roche, Guardanthealth and MSD. The authors have no other conflicts of interest to declare.
Ethical Statement: All authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Gainor JF, Dardaei L, Yoda S, et al. Molecular Mechanisms of Resistance to First- and Second-Generation ALK Inhibitors in ALK-Rearranged Lung Cancer. Cancer Discov 2016;6:1118-33. [Crossref] [PubMed]
- Recondo G, Facchinetti F, Olaussen KA, et al. Making the first move in EGFR-driven or ALK-driven NSCLC: first-generation or next-generation TKI? Nat Rev Clin Oncol 2018;15:694-708. [Crossref] [PubMed]
- Overman MJ, Modak J, Kopetz S, et al. Use of research biopsies in clinical trials: are risks and benefits adequately discussed? J Clin Oncol 2013;31:17-22. [Crossref] [PubMed]
- Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 2012;366:883-92. [Crossref] [PubMed]
- Cescon DW, Bratman SV, Chan SM, et al. Circulating tumor DNA and liquid biopsy in oncology. Nat Cancer 2020;1:276-90. [Crossref]
- Jenkins S, Yang JC, Ramalingam SS, et al. Plasma ctDNA Analysis for Detection of the EGFR T790M Mutation in Patients with Advanced Non-Small Cell Lung Cancer. J Thorac Oncol 2017;12:1061-70. [Crossref] [PubMed]
- Rolfo C, Mack PC, Scagliotti GV, et al. Liquid Biopsy for Advanced Non-Small Cell Lung Cancer (NSCLC): A Statement Paper from the IASLC. J Thorac Oncol 2018;13:1248-68. [Crossref] [PubMed]
- Sequist LV, Waltman BA, Dias-Santagata D, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med 2011;3:75ra26. [Crossref] [PubMed]
- Mehlman C, Cadranel J, Rousseau-Bussac G, et al. Resistance mechanisms to osimertinib in EGFR-mutated advanced non-small-cell lung cancer: A multicentric retrospective French study. Lung Cancer 2019;137:149-56. [Crossref] [PubMed]
- Wang Z, Yang JJ, Huang J, et al. Lung Adenocarcinoma Harboring EGFR T790M and In Trans C797S Responds to Combination Therapy of First- and Third-Generation EGFR TKIs and Shifts Allelic Configuration at Resistance. J Thorac Oncol 2017;12:1723-7. [Crossref] [PubMed]
- Hallberg B, Palmer RH. The role of the ALK receptor in cancer biology. Ann Oncol 2016;27:iii4-15. [Crossref] [PubMed]
- Solomon BJ, Besse B, Bauer TM, et al. Lorlatinib in patients with ALK-positive non-small-cell lung cancer: results from a global phase 2 study. Lancet Oncol 2018;19:1654-67. [Crossref] [PubMed]
- Zou HY, Friboulet L, Kodack DP, et al. PF-06463922, an ALK/ROS1 Inhibitor, Overcomes Resistance to First and Second Generation ALK Inhibitors in Preclinical Models. Cancer Cell 2015;28:70-81. [Crossref] [PubMed]
- Sholl LM, Aisner DL, Varella-Garcia M, et al. Multi-institutional Oncogenic Driver Mutation Analysis in Lung Adenocarcinoma: The Lung Cancer Mutation Consortium Experience. J Thorac Oncol 2015;10:768-77. [Crossref] [PubMed]
- Barlesi F, Mazieres J, Merlio JP, et al. Routine molecular profiling of patients with advanced non-small-cell lung cancer: results of a 1-year nationwide programme of the French Cooperative Thoracic Intergroup (IFCT). Lancet 2016;387:1415-26. [Crossref] [PubMed]
- Tartari CJ, Gunby RH, Coluccia AM, et al. Characterization of some molecular mechanisms governing autoactivation of the catalytic domain of the anaplastic lymphoma kinase. J Biol Chem 2008;283:3743-50. [Crossref] [PubMed]
- Hallberg B, Palmer RH. Mechanistic insight into ALK receptor tyrosine kinase in human cancer biology. Nat Rev Cancer 2013;13:685-700. [Crossref] [PubMed]
- Shinmura K, Kageyama S, Tao H, et al. EML4-ALK fusion transcripts, but no NPM-, TPM3-, CLTC-, ATIC-, or TFG-ALK fusion transcripts, in non-small cell lung carcinomas. Lung Cancer 2008;61:163-9. [Crossref] [PubMed]
- Solomon B, Varella-Garcia M, Camidge DR. ALK gene rearrangements: a new therapeutic target in a molecularly defined subset of non-small cell lung cancer. J Thorac Oncol 2009;4:1450-4. [Crossref] [PubMed]
- Desai A, Mohammed T, Rakshit S, et al. 21P The landscape of ALK alterations in non-small cell lung cancer. J Thorac Oncol 2021;16:S707. [Crossref]
- Bayliss R, Choi J, Fennell DA, et al. Molecular mechanisms that underpin EML4-ALK driven cancers and their response to targeted drugs. Cell Mol Life Sci 2016;73:1209-24. [Crossref] [PubMed]
- Lin JJ, Zhu VW, Yoda S, et al. Impact of EML4-ALK Variant on Resistance Mechanisms and Clinical Outcomes in ALK-Positive Lung Cancer. J Clin Oncol 2018;36:1199-206. [Crossref] [PubMed]
- Heuckmann JM, Balke-Want H, Malchers F, et al. Differential protein stability and ALK inhibitor sensitivity of EML4-ALK fusion variants. Clin Cancer Res 2012;18:4682-90. [Crossref] [PubMed]
- Woo CG, Seo S, Kim SW, et al. Differential protein stability and clinical responses of EML4-ALK fusion variants to various ALK inhibitors in advanced ALK-rearranged non-small cell lung cancer. Ann Oncol 2017;28:791-7. [Crossref] [PubMed]
- Camidge DR, Niu H, Kim HR, et al. Correlation of baseline molecular and clinical variables with ALK inhibitor efficacy in ALTA-1L. J Clin Oncol 2020;38:9517. [Crossref]
- Camidge DR, Dziadziuszko R, Peters S, et al. Updated Efficacy and Safety Data and Impact of the EML4-ALK Fusion Variant on the Efficacy of Alectinib in Untreated ALK-Positive Advanced Non-Small Cell Lung Cancer in the Global Phase III ALEX Study. J Thorac Oncol 2019;14:1233-43. [Crossref] [PubMed]
- Shaw AT, Kim TM, Crinò L, et al. Ceritinib versus chemotherapy in patients with ALK-rearranged non-small-cell lung cancer previously given chemotherapy and crizotinib (ASCEND-5): a randomised, controlled, open-label, phase 3 trial. Lancet Oncol 2017;18:874-86. [Crossref] [PubMed]
- Novello S, Mazières J, Oh IJ, et al. Alectinib versus chemotherapy in crizotinib-pretreated anaplastic lymphoma kinase (ALK)-positive non-small-cell lung cancer: results from the phase III ALUR study. Ann Oncol 2018;29:1409-16. [Crossref] [PubMed]
- Camidge DR, Kim HR, Ahn MJ, et al. Brigatinib versus Crizotinib in ALK-Positive Non-Small-Cell Lung Cancer. N Engl J Med 2018;379:2027-39. [Crossref] [PubMed]
- Peters S, Camidge DR, Shaw AT, et al. Alectinib versus Crizotinib in Untreated ALK-Positive Non-Small-Cell Lung Cancer. N Engl J Med 2017;377:829-38. [Crossref] [PubMed]
- Friboulet L, Li N, Katayama R, et al. The ALK inhibitor ceritinib overcomes crizotinib resistance in non-small cell lung cancer. Cancer Discov 2014;4:662-73. [Crossref] [PubMed]
- Sasaki T, Okuda K, Zheng W, et al. The neuroblastoma-associated F1174L ALK mutation causes resistance to an ALK kinase inhibitor in ALK-translocated cancers. Cancer Res 2010;70:10038-43. [Crossref] [PubMed]
- Choi YL, Soda M, Yamashita Y, et al. EML4-ALK mutations in lung cancer that confer resistance to ALK inhibitors. N Engl J Med 2010;363:1734-9. [Crossref] [PubMed]
- Katayama R, Shaw AT, Khan TM, et al. Mechanisms of acquired crizotinib resistance in ALK-rearranged lung Cancers. Sci Transl Med 2012;4:120ra17. [Crossref] [PubMed]
- Lovly CM, Pao W. Escaping ALK inhibition: mechanisms of and strategies to overcome resistance. Sci Transl Med 2012;4:120ps2. [Crossref] [PubMed]
- Heuckmann JM, Hölzel M, Sos ML, et al. ALK mutations conferring differential resistance to structurally diverse ALK inhibitors. Clin Cancer Res 2011;17:7394-401. [Crossref] [PubMed]
- Doebele RC, Pilling AB, Aisner DL, et al. Mechanisms of resistance to crizotinib in patients with ALK gene rearranged non-small cell lung cancer. Clin Cancer Res 2012;18:1472-82. [Crossref] [PubMed]
- Shaw AT, Solomon BJ, Besse B, et al. ALK Resistance Mutations and Efficacy of Lorlatinib in Advanced Anaplastic Lymphoma Kinase-Positive Non-Small-Cell Lung Cancer. J Clin Oncol 2019;37:1370-9. [Crossref] [PubMed]
- Noé J, Lovejoy A, Ou SI, et al. ALK Mutation Status Before and After Alectinib Treatment in Locally Advanced or Metastatic ALK-Positive NSCLC: Pooled Analysis of Two Prospective Trials. J Thorac Oncol 2020;15:601-8. [Crossref] [PubMed]
- Katayama R, Friboulet L, Koike S, et al. Two novel ALK mutations mediate acquired resistance to the next-generation ALK inhibitor alectinib. Clin Cancer Res 2014;20:5686-96. [Crossref] [PubMed]
- Zhang S, Anjum R, Squillace R, et al. The Potent ALK Inhibitor Brigatinib (AP26113) Overcomes Mechanisms of Resistance to First- and Second-Generation ALK Inhibitors in Preclinical Models. Clin Cancer Res 2016;22:5527-38. [Crossref] [PubMed]
- Shaw AT, Felip E, Bauer TM, et al. Lorlatinib in non-small-cell lung cancer with ALK or ROS1 rearrangement: an international, multicentre, open-label, single-arm first-in-man phase 1 trial. Lancet Oncol 2017;18:1590-9. [Crossref] [PubMed]
- Zhu VW, Lin YT, Kim DW, et al. An International Real-World Analysis of the Efficacy and Safety of Lorlatinib Through Early or Expanded Access Programs in Patients With Tyrosine Kinase Inhibitor-Refractory ALK-Positive or ROS1-Positive NSCLC. J Thorac Oncol 2020;15:1484-96. [Crossref] [PubMed]
- Shaw AT, Bauer TM, de Marinis F, et al. First-Line Lorlatinib or Crizotinib in Advanced ALK-Positive Lung Cancer. N Engl J Med 2020;383:2018-29. [Crossref] [PubMed]
- Shaw AT, Friboulet L, Leshchiner I, et al. Resensitization to Crizotinib by the Lorlatinib ALK Resistance Mutation L1198F. N Engl J Med 2016;374:54-61. [Crossref] [PubMed]
- Yoda S, Lin JJ, Lawrence MS, et al. Sequential ALK Inhibitors Can Select for Lorlatinib-Resistant Compound ALK Mutations in ALK-Positive Lung Cancer. Cancer Discov 2018;8:714-29. [Crossref] [PubMed]
- Okada K, Araki M, Sakashita T, et al. Prediction of ALK mutations mediating ALK-TKIs resistance and drug re-purposing to overcome the resistance. EBioMedicine 2019;41:105-19. [Crossref] [PubMed]
- Recondo G, Mezquita L, Facchinetti F, et al. Diverse Resistance Mechanisms to the Third-Generation ALK Inhibitor Lorlatinib in ALK-Rearranged Lung Cancer. Clin Cancer Res 2020;26:242-55. [Crossref] [PubMed]
- Takahashi K, Seto Y, Okada K, et al. Overcoming resistance by ALK compound mutation (I1171S + G1269A) after sequential treatment of multiple ALK inhibitors in non-small cell lung cancer. Thorac Cancer 2020;11:581-7. [Crossref] [PubMed]
- Lin JJ, Johnson T, Lennerz JK, et al. Resistance to lorlatinib in ROS1 fusion-positive non-small cell lung cancer. J Clin Oncol 2020;38:9611. [Crossref]
- Dagogo-Jack I, Yoda S, Lennerz JK, et al. MET Alterations Are a Recurring and Actionable Resistance Mechanism in ALK-Positive Lung Cancer. Clin Cancer Res 2020;26:2535-45. [Crossref] [PubMed]
- Sequist LV, Han JY, Ahn MJ, et al. Osimertinib plus savolitinib in patients with EGFR mutation-positive, MET-amplified, non-small-cell lung cancer after progression on EGFR tyrosine kinase inhibitors: interim results from a multicentre, open-label, phase 1b study. Lancet Oncol 2020;21:373-86. [Crossref] [PubMed]
- Coleman N, Wotherspoon A, Yousaf N, et al. Transformation to neuroendocrine carcinoma as a resistance mechanism to lorlatinib. Lung Cancer 2019;134:117-20. [Crossref] [PubMed]
- Mezquita L, Swalduz A, Jovelet C, et al. Clinical Relevance of an Amplicon-Based Liquid Biopsy for Detecting ALK and ROS1 Fusion and Resistance Mutations in Patients With Non-Small-Cell Lung Cancer. JCO Precis Oncol 2020;4:PO.19.00281.
- Horn L, Whisenant JG, Wakelee H, et al. Monitoring Therapeutic Response and Resistance: Analysis of Circulating Tumor DNA in Patients With ALK+ Lung Cancer. J Thorac Oncol 2019;14:1901-11. [Crossref] [PubMed]
- Dagogo-Jack I, Rooney M, Lin JJ, et al. Treatment with Next-Generation ALK Inhibitors Fuels Plasma ALK Mutation Diversity. Clin Cancer Res 2019;25:6662-70. [Crossref] [PubMed]
- Pailler E, Faugeroux V, Oulhen M, et al. Acquired Resistance Mutations to ALK Inhibitors Identified by Single Circulating Tumor Cell Sequencing in ALK-Rearranged Non-Small-Cell Lung Cancer. Clin Cancer Res 2019;25:6671-82. [Crossref] [PubMed]
- Cui JJ, Rogers E, Zhai D, et al. Abstract 5226: TPX-0131: A next generation macrocyclic ALK inhibitor that overcomes ALK resistant mutations refractory to current approved ALK inhibitors. Cancer Res 2020;80:5226.
- Odegaard JI, Vincent JJ, Mortimer S, et al. Validation of a Plasma-Based Comprehensive Cancer Genotyping Assay Utilizing Orthogonal Tissue- and Plasma-Based Methodologies. Clin Cancer Res 2018;24:3539-49. [Crossref] [PubMed]
Cite this article as: Blaquier JB, Cardona AF, Russo A, Rolfo C, Recondo G. Next-generation sequencing using liquid biopsy in the care of patients with ALK-rearranged non-small cell lung cancer: a focus on lorlatinib. Precis Cancer Med 2021;4:28.

