
Non-small cell lung cancer molecular characterization of advanced disease with focus on sex differences: a narrative review
Introduction
Lung cancer is the leading cause of cancer related mortality worldwide (11% of all cancers) with a prevalence of advanced stage in up to 70% of cases (1,2). In particular, lung cancer shows the highest incidence and mortality in males, and ranks third in incidence and second in mortality among females (2).
There are two main histological types of lung cancer: small cell lung cancer (SCLC), accounting for 15% of cases, derives from cells with neuroendocrine characteristics and has a highly malignant behaviour, and the more common non-small cell lung cancer (NSCLC), accounting for 85% of cases (3,4). NSCLC is further divided into the following subtypes: adenocarcinoma (ADC), which is the most frequent (40%); squamous cell carcinoma (SCC) (25–30%); and large cell carcinoma (5–10%) (4).
NSCLC subtype discrimination drives molecular analysis in the field of precision medicine. Molecular characterization is crucial for advanced lung adenocarcinoma patients, who can benefit from targeted therapies in presence of actionable gene alterations. According to the national and international guidelines, all patients with advanced adenocarcinoma—regardless of clinical indicators such as smoking status, race or sex—should undergo molecular testing for at least epidermal growth factor receptor (EGFR), ALK receptor tyrosine kinase (ALK), ROS proto-oncogene 1, receptor tyrosine kinase (ROS1), ret proto-oncogene (RET), B-Raf proto-oncogene, serine/threonine kinase (BRAF), and neurotrophic receptor tyrosine kinase (NTRK), whose alterations are predictive of response to approved kinase inhibitors. Furthermore, programmed cell death ligand 1 (PD-L1) evaluation in both ADC and SCC is necessary to select patients for immunotherapy (5,6). Other biomarkers are currently under investigation and their analysis is recommended whenever possible (6,7). Despite the initial response to targeted therapies, the majority of patients develop resistance within 1 year; consequently, alongside the identification of targetable alterations, an accurate molecular description of resistance mechanisms can improve patient management, leading to other treatment regimens (8,9).
It has been demonstrated that the evaluation of co-occurring mutations and the study of tumor heterogeneity are crucial to better understand the tumor molecular landscape, which can differently affect the response to therapies (10). In this respect, peculiar molecular characteristics and a different incidence of driver alterations mainly related to tobacco exposure and hormone regulation have been reported between males and females in lung cancer. Although not fully understood and described, sex-related molecular features can have important consequences on prognosis and response to therapy (11-15).
The aim of this paper is to review the most important predictive biomarkers in advanced NSCLC as well as the current knowledge about molecular differences between men and women with a brief reference to our seven-year single centre experience in histological and molecular diagnosis of lung cancer.
We present the following article in accordance with the Narrative Review Reporting Checklist (available at: http://dx.doi.org/10.21037/pcm-20-72).
Targeted therapies
Targeted therapies in lung ADC rely on the use of kinase inhibitors able to target driver oncoproteins, whose signaling pathways promote tumor survival and proliferation. Targetable alterations mainly occur in genes encoding for the tyrosine kinase receptors such as EGFR, MET proto-oncogene, receptor tyrosine kinase (MET), erb-b2 receptor tyrosine kinase 2 (HER2), ALK, ROS1, RET, NTRK, and for cytoplasmatic proteins with kinase activity including KRAS proto-oncogene, GTPase (KRAS) and BRAF (6,16).
EGFR
EGFR is the main actionable target in advanced lung adenocarcinoma, with a mutation frequency of 15% and 40% in Caucasian and Asiatic patients, respectively (17,18). Patients with advanced lung adenocarcinoma harboring EGFR activating mutations can achieve greater benefit from treatment with tyrosine kinase inhibitors (TKIs) than with platinum standard chemotherapy (19). EGFR mutations responsible for the constitutive activation of the protein involve exons 18, 19, 20 and 21, encoding for the tyrosine kinase (TK) domain. About 90% of cases are in frame deletions within exon 19 or the missense mutation L858R in the exon 21 (19,20).
Five TKIs have been approved for the first-line treatment of EGFR mutant lung ADC: the first generation TKIs erlotinib and gefitinib, which reversibly bind to the TK domain (21-23); the second generation irreversible TKIs afatinib and dacomitinib, which also inhibit HER2 (24-27); the third generation irreversible and mutant-selective osimertinib, active against both the sensitive mutations and the main resistance mutation T790M in the exon 20 (28,29).
Almost all patients treated with EGFR TKIs develop resistance within 1 year. The most important resistance mechanism to first and second generation TKIs is the secondary EGFR mutation T790M, which increases affinity for ATP (30). The presence of T790M, detected both in tumor tissue and in tumor circulating cell-free DNA (ctDNA), makes patients eligible for second-line treatment with osimertinib (28). Other significant resistance mechanisms are HER2 amplifications (more common for first generation TKIs), MET amplifications, histological transformation in SCLC, and mutations in BRAF, KRAS and phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha (PIK3CA) genes (31).
Resistance mechanisms to osimertinib are more heterogenous and differ between first and second line settings. After second-line osimertinib treatment, in 10–26% of cases a tertiary EGFR mutation is detected within exon 20, the C797S, with or without a co-occurring T790M. Whenever C797S co-occurs with T790M, the definition of the allelic conformation (cis or trans) can drive the following line of treatment. Indeed, the co-occurrence of these two resistance mutations on the same allele (cis) leads to a mutant protein that cannot be targeted by currently approved TKIs. Otherwise, a TKI combination therapy can be suggested (32). At progression time to second-line osimertinib, about 49% of cases can lose T790M and also MET amplification and histological transformation are frequently observed (9). After first-line osimertinib treatment, T790M is not detected, MET amplification and histological transformation are among the main resistance mechanisms, and the frequency of EGFR secondary mutations, like C797S, is about 10–15% (9,33).
It has been reported that several resistance mechanisms can co-occur in the same tumor, thus exacerbating the tumor heterogeneity issue. Consequently, a multi-gene analysis of both tissue re-biopsy and ctDNA is advisable (9,34).
Apart from the most frequent targetable alterations, about 10–14% of EGFR-mutated lung cancers harbor uncommon mutations alone or co-occurring with the most common activating mutations (i.e., exon 19 deletions and L858R) (35). The main sensitive rare mutations are E709x (1.5%) and G719x (1.5–3%) in exon 18, S768I (0.6–1%) in exon 20 and L861Q (3%) in exon 21 (20); patients harboring these mutations can be treated with afatinib (36). On the other hand, exon 20 insertions, which have an incidence of 4–10% among EGFR mutations, are well-known de novo resistance alterations, except for the insertion A763_Y764insFQEA, which has shown sensitivity to EGFR TKIs (37). Exon 20 insertions, for a long time considered untargetable, are currently under investigation with allosteric compounds, some of which (e.g., poziotinib), have already shown promising results, with a reported response rate of 64% in a phase-II clinical trial (38,39).
BRAF
BRAF encodes for a serine threonine kinase and is mutated in 5–8% of NSCLC, with a higher prevalence in lung ADC (40,41).
In advanced lung ADC, the V600E mutation, accounting for 50% of all BRAF mutated cases, is a predictive biomarker (42) designed to select patients for front-line treatment with dabrafenib (a BRAF inhibitor), together with trametinib (a MEK inhibitor) (43). Resistance mechanisms to BRAF and MEK TKIs have not been clearly described in lung cancer, but the principal ones include MEK activating alterations, RAS mutations, phosphatase and tensin homolog (PTEN) inactivation, as well as tyrosine kinase receptors overexpression (44).
BRAF mutations have also been described as resistance mechanisms to some TKI treatments as those targeting EGFR or ROS1 (31,45).
Finally, it is worth noting that BRAF V600E mutation in lung cancer as well as targetable alterations in EGFR, ROS1 and ALK, are associated with an immunosuppressive tumor microenvironment, whereas non V600E mutations (i.e., K601E, L597Q, G469A), more frequently detected in smokers, are linked to an immunogenic tumor microenvironment. This aspect can have implications in patient selection for immune check-point inhibitors (46).
MET-HER2-KRAS
MET alterations have been described in advanced lung ADC as both primary oncogenic drivers and drivers of acquired resistance to EGFR TKIs (47). MET amplification and alterations causing the skipping of exon 14 can be targeted. More specifically, MET amplification is reported in 2–5% of NSCLC (48), and its incidence is higher after treatment with EGFR TKI (9,31). Combination therapies including EGFR and MET inhibitors are currently under investigations (47). MET exon 14 skipping alterations are reported in 2–4% of NSCLC and are the most common alterations in sarcomatoid carcinomas (49,50). Interestingly, in 5% of cases MET mutations can co-occur with other driver alterations mainly in RAS genes (51). The multi-kinases inhibitors crizotinib and cabozantinib have been approved by FDA as a breakthrough-therapy designation for the treatment of patients who progressed after receiving platinum-based chemotherapy, and capmatinib was approved also for first-line setting (52). The MET-selective inhibitors capmatinib, savolitinib, and tepotinib also showed valuable results especially when used in combination with anti-EGFR TKIs (53,54). Tepotinib was approved by the FDA with a breakthrough-therapy designation of patients—affected by metastatic NSCLC harboring MET exon 14 skipping alterations—who progressed after platinum-based chemotherapy (55).
HER2 is a tyrosine kinase receptor from the same family of EGFR. Its main alterations in lung ADC are exon 20 insertions (2–9% of cases) (56) and amplifications, with the latter mainly described as EGFR TKI resistance mechanisms (57). The targeted drugs currently available have a limited activity in HER2-mutant NSCLC, but none of these have been approved so far (58). However, poziotinib has shown promising preclinical and early clinical activity in NSCLC patients with HER2 or EGFR exon 20 insertions (39,59).
KRAS encodes for a GTPase protein and is mutated in 20–30% of lung ADC patients, mainly smokers, with G12C being the most frequent alteration (40%) (7). Over the years, several attempts have been made to target this key protein for tumor progression, but promising results have been obtained only recently with the development of KRAS G12C specific inhibitors, among which AMG159 and MRTX849 are currently investigated (60).
Gene fusions
The ALK gene is rearranged in 3% to 5% of NSCLC cases (61), typically occurring in younger, in never- or light-smokers, and in adenocarcinoma patients (62). The most frequent ALK rearrangements are caused by pericentric inversions of the short arm of chromosome 2, leading to a fusion gene between the amino terminal portion of the Echinoderm Microtubule-associated protein Like 4 (EML-4) gene and the juxtamembrane region of the ALK gene (63). As well as EML4, more than 90 other ALK partners have been reported in NSCLC, including the kinesin family member 5B (KIF5B), trafficking from ER to golgi regulator (TFG), the kinesin light chain 1 (KLC1), and the huntingtin interacting protein 1 (HIPI) (64,65). The multi-kinases inhibitor crizotinib (first generation ALK TKI) is the first to have been approved to treat ALK-positive lung cancer patients (66,67). Despite its initial significant benefit, the efficacy of crizotinib decreases after the onset of acquired resistance mechanisms, mainly secondary mutations in the ALK gene and activation of bypass tracks (68). Several innovative ALK inhibitors have been approved in both crizotinib-naïve and resistant ALK-rearranged NSCLC including ceritinib, alectinib, brigatinib and lorlatinib. At present, alectinib is also the preferred agent for first-line treatment (69).
ROS1 is activated by chromosomal rearrangement in about 1% of NSCLC cases (70,71). Well-known ROS1 fusion partners in lung cancer include the CD74 molecule (CD74), which is the most frequent, DEPP1 autophagy regulator (DEPP1), the solute carrier family 34 member 2 (SLC34A2), and syndecan 4 (SDC4). The kinase domains of ALK and ROS1 share 77% of amino acid identity within the ATP-binding sites and can be targeted by the same inhibitors, including crizotinib (67). Ceritinib, lorlatinib (72,73), entrectinib (74), and repotrectinib have also proved to be safe and active in patients with advanced ROS1-rearranged NSCLC (75).
RET gain-of-function mutations or rearrangements occurs in 1–2% of NSCLC patients (76). RET rearrangements involve at least 12 fusion partners, the most common being the kinesin family 5B (KIF5B) gene, followed by the coiled-coil domain containing 6 (CCDC6) gene, the nuclear receptor coactivator 4 (NCOA4) gene or the tripartite motif containing 33 (TRIM33) gene (77). RET fusions are mutually exclusive with other driver mutations and more commonly appear in younger patients and light-/never-smokers (78). Selpercatinib has recently been approved for the first-line treatment of RET-rearranged NSCLC according to the results of the LIBRETTO-001 trial (79). In addition, cabozantinib proved to be effective as second-line treatment with a median progression free survival (PFS) equal to 5.5 months and a median overall survival (OS) of 9.9 months in 25 patients included in a phase II-trial (80).
The tropomyosin receptor kinase (TRK) family consists of three tyrosine kinase receptors: TRKA, TRKB and TRKC isoforms, encoded by the NTRK1, NTRK2 and NTRK3 genes, respectively. They are predominantly expressed in the nervous system, where they act as important modulators in the development of neuronal and other tissues (e.g., lung, bone) (81). NTRK fusions involve over 80 partner genes that promote constitutive TRK signaling activation, cell transformation, and proliferation (82,83). NTRK fusions have a prevalence of 0.1–1% in unselected NSCLC, which reaches about 3% in tumors lacking EGFR, KRAS, ALK and ROS1 alterations (81,82). In the last two years, FDA has granted accelerated approval to the two first-generation selective TRK inhibitors, larotrectinib (84) and entrectinib as tumor-agnostic treatment after the impressive clinical activity in patients whose tumors harbored NTRK fusions (85). Future clinical research in the setting of these tumors is directed towards overcoming resistance to first-generation TRK inhibitors.
Immunotherapy
Immunotherapy aims to induce or enhance the cancer-specific immune response and, in this context, the use of check-point inhibitors has been approved as first- or second-line treatments in different solid tumors. PD-L1/PD-1 is the most important immune checkpoint, associated with immune tolerance and autoimmune diseases, and is used by the tumor to elude the immune system. PD-1 is encoded by the programmed cell death 1 (PDCD1) gene, and it is expressed in immune cells such as T-cells, B-cells, natural killer cells, and dendritic cells. PD-L1 was found to be not only confined to the surface of tumor cells, but also expressed on the surface of antigen-presenting cells (APC), Treg cells, infiltrating myeloid cells, and others. When binding to its ligands, PD-1 can inhibit the activation of lymphocytes and induce the death of lymphocytes (86,87). Therefore, agents targeting the PD-L1/PD-1 axis have become a hotspot of anti-tumor treatments. The PD-L1 protein expression detected by immunohistochemistry (IHC) is the predictive biomarker used in most immuno-oncology clinical trials (88).
Anti PD-1 pembrolizumab and nivolumab and anti PD-L1 atezolizumab are approved by FDA as monotherapies for advanced NSCLC. Based on the outcomes of a series of clinical trials, PD-L1 testing has been recommended to select patients for Pembrolizumab monotherapy as first-line treatment (PD-L1 ≥50%) (89-92). The predictive value of PD-L1 expression has proven to be quite satisfactory for first-line pembrolizumab/atezolizumab monotherapy, but it is not ideal to select patients for pembrolizumab-based combination regimens as first-line treatments [Keynote-189 and Keynote-407, (93,94)]. In the second-line setting, Checkmate-017 and Checkmate-057 studies have demonstrated superior OS of nivolumab monotherapy over docetaxel, regardless of the PD-L1 expression (95,96). However, POPLAR and OAK have reported that only patients with a PD-L1 expression on tumor cells or on immune cells greater than 1% could benefit from immunotherapy, while patients with expression levels below 1% did not achieve a better OS compared to docetaxel (97,98). Variable PD-L1 predictive values have been reported across different clinical trials testing anti PD-1 or PD-L1 agents, and different studies have shown the imperfect predictive value of this biomarker.
In this respect, a better determination of tumor antigenicity and tumor microenvironment can help to identify patients eligible for immunotherapy. Tumor mutational burden (TMB) is related to the number of mutations per megabase. A high number of somatic mutations may lead to a greater number of neoantigens presented on the surface of tumor cells, which, in turn, may increase immunogenicity (99). The relation between a higher mutational burden and a better response to checkpoint inhibition has been demonstrated in several studies. In particular, the post-hoc analysis of Checkmate 227 demonstrated that a higher TMB is associated with prolonged PFS independent of PD-L1 expression and histology in patients receiving first-line nivolumab plus ipilimumab in a metastatic setting. However, OS revealed no benefit related to high TMB (100). In spite of its potentiality, TMB is not a biomarker easy to evaluate in clinical practice; in addition, a standardization procedure and a univocal cut-off are still missing.
Immune gene signatures have been also evaluated to characterize the tumor microenviroment. In particular IFN-γ signaling and activated T-cells have shown a good predictive value in identifying responders to immunotherapy across several solid tumors including melanoma and NSCLC (101).
Molecular tests
The increasing number of predictive biomarkers in lung cancer and the development of new drugs make it necessary to perform a complete evaluation of tumor molecular status. However, molecular characterization of advanced lung cancer can be hampered by the availability of biological material. As a matter of fact, most lung cancer patients are diagnosed at advanced stage, and in about 50% of cases only cytology is available (102).
Compared to other molecular tests, the use of next generation sequencing (NGS) multi-gene panels is recommended, since NGS panels allow to evaluate several markers simultaneously by using a few biological materials. NGS is the ideal method to analyse also rare mutations seldom included in hotspot tests, thus providing additional information that is relevant to the clinical context (35,103). Moreover, the use of NGS is necessary to explore tumor heterogeneity and to identify co-occurring mutations with an important impact on prognosis and response to therapies (10,104). NGS panels are also an advantage for the analysis of ctDNA, which is essential to monitor the response to treatment and to evaluate resistance mechanisms (105).
Different diagnostic algorithms regarding gene fusions are currently used, mainly based on fluorescent in situ hybridization (FISH) and IHC. The detection of the ALK fusion protein by IHC is the gold standard to select patients eligible for ALK TKI treatment. However, moderate or weak ALK immunostaining requires FISH confirmation (106). ROS1 rearrangements can be detected by FISH assays using a dual-color break-apart probe, but the detection of elevated ROS1 protein levels by IHC may provide cost-effective screening, considering the rarity of these rearrangements in NSCLC. Confirmation of positive or doubtful ROS1 immunostaining by FISH or by other methods is highly recommended (106). FISH and sequencing techniques are considered the standard methods to detect RET rearrangements, considering the low accuracy demonstrated by IHC (76). NGS is the most commonly used and specific method to detect NTRK fusions (107). IHC can detect TRK proteins overexpression, which may reflect the presence of NTRK fusions. This approach has shown an overall sensitivity of 88%, which is lower for NTRK3 fusions (79%) than NTRK1 and NTRK2 (about 97%) (107). IHC can therefore be used in clinical practice as a screening method to identify NTRK fusions followed by a confirmatory NGS test. Despite the cost-effectiveness of FISH and IHC, the evidence that specific fusion variants can have a different sensitivity to TKIs (108) has favored the development and validation of several multi-marker panels running on both NGS and non-NGS platforms (109-111).
Although there is a lack of standardization and expertise for a correct interpretation of the results, multi-marker NGS panels are currently the main protagonists of lung cancer molecular characterization, with different panels already validated and approved for clinical practice (112).
Sex impact on the molecular landscape of NSCLC
Sex differences can impact on incidence, prognosis, mortality and response to therapies (14). To date, the molecular basis for sex disparities is still to be fully understood, but it has been demonstrated that men have a higher mortality rate than women, who better respond to both surgery and chemotherapy (113).
Sex differences are mainly due to smoking habits and hormonal status (114,115). Epidemiological studies have demonstrated that lung cancer in women is less associated with tobacco exposure. In fact, lung cancer in never smokers has an incidence of 10–15%, and is more frequent in women (53%) than in men (15%) (114). Among others, an increased risk of lung cancer in women can also be correlated to biomass fuel and cooking and to a higher susceptibility to chronic obstructive pulmonary disease than in men (114).
Discordant data have been reported concerning different risks of developing smoking-related lung cancer in males and females (114,116). However, some differences in the carcinogenic effects of tobacco have been identified: in women, cytochrome P450 family 1 subfamily A member 1 (CYP1A1) is highly expressed, and glutathione S transferase M1 (GSTM1) has a higher rate of mutations or polymorphisms that decrease its activity (117). CYP1A1 activates polycyclic aromatic hydrocarbons to highly reactive compounds capable of binding DNA, whereas GSTM1 detoxifies active forms of polycyclic aromatic hydrocarbons (117). Accordingly, female smokers have higher levels of DNA adducts compared to male smokers. Tumor protein p53 (TP53) mutations have a higher frequency in smoking women than in smoking men, whereas the opposite is observed in non-smokers (118). Finally, a lower efficiency of the DNA repair system has been revealed in both smoker and non-smoker women (119-121). Adenocarcinoma is the most frequent histotype of NSCLC, with a higher prevalence in women, who more often present with driver alterations in targetable oncogenes. EGFR mutations and BRAF V600E are more frequent in non-smoker females than in non-smoker men (114,122). Similarly, HER2 mutations and MET exon 14 skipping mutations are associated with female sex, never-smoked status and adenocarcinoma histology (51,123). On the contrary, KRAS mutations are more common in smokers, with G12C being the most frequent alteration, especially in women (124). Comprehensive molecular profiling of lung ADC performed on tumor and paired normal tissues from 230 untreated lung ADC patients revealed that only a fraction of significantly mutated genes is enriched in men or women. Among these genes, EGFR mutations were more frequent in females, whereas loss of function mutations in the gene RNA binding motif protein 10 (RBM10), located on chromosome X, were more common in men (125). Other X-linked genes were confirmed to play an important role in the biology of lung cancer. These genes included the Gastrin-peptide growth factor receptor (GRPR), which is more commonly expressed in women than in men both smokers and non-smokers. This receptor enhances cell proliferation and metastases and can be regulated by estrogens (126).
Metabolic abnormalities in lung cancer differ between sexes, as demonstrated by Li and collaborators who performed a transcriptome analysis using The Cancer Genome Atlas (TCGA) data. In particular, they identified metabolic genes differently expressed and impacting on prognosis in a sex-biased manner. TAO kinase 2 (TAOK2), a serine/threonine protein kinase catalytically activated during mitosis, was found to influence male prognosis. On the other hand, N-acylsphingosine amidohydrolase 1 (ASAH1), which catalyzes the hydrolysis of ceramide into sphingosine and may act as an oncogene, seems to have a critical role in women. The authors identified a total of 34 and 15 metabolic genes in men and women respectively, as potential diagnostic and prognostic sex-biased biomarkers for lung ADC (127).
Hormonal status can explain some sex differences in human tumors considering that estrogens are crucial for cancer development (128). For instance, 17-b-estradiol impacts on lung cancer and can activate the EGFR pathway (129). The estrogen receptors (ER) expressed in the lung epithelium are ER-alpha, which allows a proper differentiation of the lung and an adequate number of alveoli per surface, and ER-beta, which is involved in the development of the extracellular matrix (130). ER-beta is associated with EGFR mutations (131), and is highly expressed in lung cancer, especially in premenopausal women. Consistently, the inhibition of aromatase, crucial for estrogen synthesis, may improve the response to EGFR TKIs (129).
Cheng and collaborators examined the expression of the hormone receptor in lung tumors, finding that women had a lower cytoplasmatic ER-alpha and nuclear ER-beta expression than men. Higher cytoplasmatic ER-alpha and nuclear ER-beta expression is associated with a worse survival (132).
Dubois evaluated the contribution of lymphatic and blood endothelial cells in the sex-dependent modulation of lung cancer. They discovered that lung tumors had a faster growth in female than in male mice, and that estradiol specifically promoted tumor development in females. They observed that estradiol increased lymph-angiogenesis and the levels of the vascular endothelial growth factor A (VEGFA) and of the fibroblast growth factor 2 (bFGF) through the ER-alpha pathway. They also reported that the use of tamoxifen and of ER-alpha antagonist decreased lung tumor volume, altered blood and lymphatic vasculature, and reduced VEGFA and bFGF levels only in females (133).
Cancer in males and females also present some endocrine differences. Parathyroid hormone-related protein (PTHrP) is expressed in NSCLC and is upregulated in tumors presenting skeletal metastasis. It has been demonstrated that the PTHrP expression has a positive impact on OS in females, whereas survival in males is independent of PTHrP (134,135).
Sex differences were also reported in both innate and adaptive immune responses, with more antigenic tumors in men. Immune checkpoint inhibitors tend to be more effective in male patients, while immunotherapy combined with chemotherapy is more effective in females. These sex differences may again result from hormonal factors and X-linked genes (11,136). Immune-related adverse events are more frequent in women, especially pneumonitis and endocrinopaties, but a better PFS was observed in women with adverse events (136). Caetano et al demonstrated a sex-specific role for epithelial STAT3 signaling in the KRAS-mutant mouse model of lung adenocarcinoma. Specifically, the absence of epithelial STAT3 in males favours lung tumorigenesis through IL-6 signaling and neutrophilic inflammation, which is diminished by estrogen/ER signaling in females (137).
A recent meta-analysis has evaluated the impact of sex and age on the treatment of lung cancer by analysing data from representative Phase III-clinical trials and also by evaluating TKI efficacy. The authors reported sex differences related to treatment efficacy: women respond better whenever the drug shows activity, and this suggests that treatment should be evaluated also considering sex (138). Likewise, Buonerba and collaborators found that male smoker patients positive for L858R EGFR mutation may benefit less from EGFR TKI treatment (17).
Yuan et al. reported that in the lung adenocarcinoma cohort of TCGA, the serine/threonine kinase 11 (STK11) gene is more frequently mutated in males; and that STK11 inactivating mutations may predict sensitivity to mTOR and SRC inhibitors (139). On the other hand, neurofibromin 1 (NF1) in SCC is more frequently deleted in women and its inactivation is associated with mTOR and MEK inhibitors sensitivity (139-141).
Finally, Radkiewicz and collaborators reported that sex differences are less evident in SCC than in ADC and they confirmed a different rate of EGFR mutations (15).
Sex differences in lung cancer: a single centre seven-year experience
The availability of large databases and NGS techniques has increased our knowledge of sex differences in lung cancer, but only a few data are available regarding differences in targetable alterations between Caucasian men and women with advanced lung cancer. In this paper, we report data from our single centre seven-year experience including 2,425 consecutive patients diagnosed with advanced lung cancer, who underwent molecular characterization at the Unit of Pathological Anatomy of the University Hospital of Pisa. In detail, 1,514 men (median age 68.2±10.7 years) and 911 women (median age 65.4±9.4 years) were tested for the mutational status of EGFR, BRAF, KRAS, MET and PIK3CA; in addition, ALK, RET and ROS1 fusions, as well as HER2 and MET amplifications were also evaluated. Figure 1 shows the histological and molecular data.
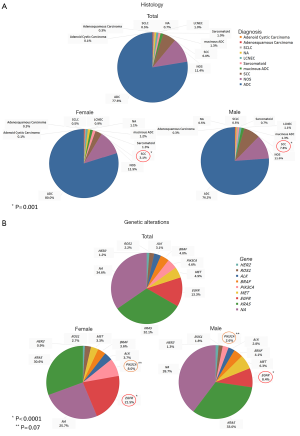
The SCC histotype was significantly more frequent in men, while the EGFR mutations were more common in women. Similarly to EGFR, PIK3CA alterations tended to be more frequent in women. No differences were observed for the other biomarkers. Interestingly, exon 19 EGFR deletions were significantly more frequent in women, whereas L858R was more frequent in men (Figure 2).
The rate of EGFR mutation types associated with sex has not been clearly assessed, but it has been demonstrated that exon 19 deletions usually occur in younger patients (142), and, in our cohort, females were significantly younger than men (P<0.001). Koyama and collaborators found that patients with tumors harboring an exon 19 EGFR deletion have better OS compared to L858R after treatment with EGFR TKI (143), thus supporting the overall better response to TKIs in women. As regards the TKI resistance mechanisms, no differences were observed between tumors harboring exon 19 deletions and L858R (144). PIK3CA mutations have a frequency of 2% to 5% in NSCLC and are usually more prevalent in SCC (145). In adenocarcinoma, PIK3CA alterations may indicate a worse prognosis and have also been described as concurrent with other oncogenic drivers like EGFR, impacting on sensitivity to TKIs (146).
Conclusions
Precision medicine has greatly improved the management of patients with advanced lung adenocarcinomas (1). In this context, molecular characterization is crucial to assess the best therapeutic options both in first and further line settings (6). Several predictive biomarkers have been introduced in clinical practice, and there is still an increase in the number of variants that should be routinely characterized (16).
Many targetable alterations in oncogenes have well known clinical implications, but some aspects related to the molecular landscape of lung cancer need to be better clarified. It has been demonstrated that the single oncogenic driver paradigm is not completely appropriate within the context of lung cancer. Tumor heterogeneity and co-occurring alterations, especially in oncosuppressor genes like TP53 or STK11, can define subgroups among lung cancers addicted to the same oncogene, with different responses to therapies and tumor microenvironment (10). The identification of resistance mechanisms can be challenging, and the selection of patients eligible for immunotherapy is still a debated topic. In this regard, sex differences contribute to peculiar molecular characteristics. Molecular differences between sexes are mainly due to tobacco exposure and hormonal status and lead to different frequencies of driver alterations, response to therapy and prognosis (114,115). A higher incidence of targetable driver alterations has been reported in non-smoker women compared to men, and sex-specific alterations in genes involved in tobacco metabolism have been identified (114). Moreover, hormonal status influences cancer development and progression and can be associated with the activation of targetable pathways. Different responses to TKIs between men and women harboring the same driver alterations have been highlighted; susceptibility to immunotherapy is also different between sexes (11,17).
Although interesting data are already available, the basis of molecular differences between sexes is not fully understood and further studies are necessary to evaluate the predictive values of sex-related molecular features in clinical practice.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Editta Baldini and Franca Melfi) for the series “Lung Cancer In Women: From Epidemiology To Therapy” published in Precision Cancer Medicine. The article has undergone external peer review.
Reporting Checklist: The authors have completed the Narrative Review Reporting Checklist. Available at http://dx.doi.org/10.21037/pcm-20-72
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/pcm-20-72). The series “Lung Cancer In Women: From Epidemiology To Therapy” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Domagala-Kulawik J. New Frontiers for Molecular Pathology. Front Med (Lausanne) 2019;6:284. [Crossref] [PubMed]
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Rotow J, Bivona TG. Understanding and targeting resistance mechanisms in NSCLC. Nat Rev Cancer 2017;17:637-58. [Crossref] [PubMed]
- Travis WD, Brambilla E, Burke AP, et al. WHO classification of tumours of lung, pleura, thymus and heart. 4th edition. Lyon 2015. IARC Publications.
- Pennell NA, Arcila ME, Gandara DR, et al. Biomarker Testing for Patients With Advanced Non-Small Cell Lung Cancer: Real-World Issues and Tough Choices. Am Soc Clin Oncol Educ Book 2019;39:531-42. [Crossref] [PubMed]
- Lindeman NI, Cagle PT, Aisner DL, et al. Updated Molecular Testing Guideline for the Selection of Lung Cancer Patients for Treatment With Targeted Tyrosine Kinase Inhibitors: Guideline From the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. Arch Pathol Lab Med 2018;142:321-46. [Crossref] [PubMed]
- Christensen JG, Olson P, Briere T, et al. Targeting Kras g12c‐mutant cancer with a mutation‐specific inhibitor. J Intern Med 2020;288:183-91. [Crossref] [PubMed]
- Liu WJ, Du Y, Wen R, et al. Drug resistance to targeted therapeutic strategies in non-small cell lung cancer. Pharmacol Ther 2020;206:107438. [Crossref] [PubMed]
- Leonetti A, Sharma S, Minari R, et al. Resistance mechanisms to osimertinib in EGFR-mutated non-small cell lung cancer. Br J Cancer 2019;121:725-37. [Crossref] [PubMed]
- Skoulidis F, Heymach JV. Co-occurring genomic alterations in non-small-cell lung cancer biology and therapy. Nat Rev Cancer 2019;19:495-509. [Crossref] [PubMed]
- Wang S, Cowley LA, Liu XS. Sex Differences in Cancer Immunotherapy Efficacy, Biomarkers, and Therapeutic Strategy. Molecules 2019;24:3214. [Crossref] [PubMed]
- Wakelee HA, Chang ET, Gomez SL, et al. Lung Cancer Incidence in Never Smokers. J Clin Oncol 2007;25:472-8. [Crossref] [PubMed]
- Kim HI, Lim H, Moon A. Sex Differences in Cancer: Epidemiology, Genetics and Therapy. Biomol Ther (Seoul) 2018;26:335-42. [Crossref] [PubMed]
- Li CH, Haider S, Shiah YJ, et al. Sex Differences in Cancer Driver Genes and Biomarkers. Cancer Res 2018;78:5527-37. [Crossref] [PubMed]
- Radkiewicz C, Dickman PW, Johansson ALV, et al. Sex and survival in non-small cell lung cancer: A nationwide cohort study. PLoS One 2019;14:e0219206. [Crossref] [PubMed]
- Yang SR, Schultheis AM, Yu H, et al. Precision medicine in non-small cell lung cancer: Current applications and future directions. Semin Cancer Biol 2020; [Crossref] [PubMed]
- Buonerba C, Iaccarino S, Dolce P, et al. Predictors of Outcomes in Patients with EGFR-Mutated Non-Small Cell Lung Cancer Receiving EGFR Tyrosine Kinase Inhibitors: A Systematic Review and Meta-Analysis. Cancers (Basel) 2019;11:1259. [Crossref] [PubMed]
- Shi Y, Au JS, Thongprasert S, et al. A Prospective, Molecular Epidemiology Study of EGFR Mutations in Asian Patients with Advanced Non-Small-Cell Lung Cancer of Adenocarcinoma Histology (PIONEER). J Thorac Oncol 2014;9:154-62. [Crossref] [PubMed]
- Aran V, Omerovic J. Current Approaches in NSCLC Targeting K-RAS and EGFR. Int J Mol Sci 2019;20:5701. [Crossref] [PubMed]
- Zhang T, Wan B, Zhao Y, et al. Treatment of uncommon EGFR mutations in non-small cell lung cancer: new evidence and treatment. Transl Lung Cancer Res 2019;8:302-16. [Crossref] [PubMed]
- Mitsudomi T, Morita S, Yatabe Y, et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol 2010;11:121-8. [Crossref] [PubMed]
- Zhou C, Wu YL, Chen G, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol 2011;12:735-42. [Crossref] [PubMed]
- Rosell R, Carcereny E, Gervais R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol 2012;13:239-46. [Crossref] [PubMed]
- Sequist LV, Yang JC, Yamamoto N, et al. Phase III Study of Afatinib or Cisplatin Plus Pemetrexed in Patients With Metastatic Lung Adenocarcinoma With EGFR Mutations. J Clin Oncol 2013;31:3327-34. [Crossref] [PubMed]
- Wu YL, Zhou C, Hu CP, et al. Afatinib versus cisplatin plus gemcitabine for first-line treatment of Asian patients with advanced non-small-cell lung cancer harbouring EGFR mutations (LUX-Lung 6): an open-label, randomised phase 3 trial. Lancet Oncol 2014;15:213-22. [Crossref] [PubMed]
- Park K, Tan EH, O’Byrne K, et al. Afatinib versus gefitinib as first-line treatment of patients with EGFR mutation-positive non-small-cell lung cancer (LUX-Lung 7): a phase 2B, open-label, randomised controlled trial. Lancet Oncol 2016;17:577-89. [Crossref] [PubMed]
- Wu YL, Cheng Y, Zhou X, et al. Dacomitinib versus gefitinib as first-line treatment for patients with EGFR-mutation-positive non-small-cell lung cancer (ARCHER 1050): a randomised, open-label, phase 3 trial. Lancet Oncol 2017;18:1454-66. [Crossref] [PubMed]
- Mok TS, Wu YL, Ahn MJ, et al. Osimertinib or Platinum-Pemetrexed in EGFR T790M-Positive Lung Cancer. N Engl J Med 2017;376:629-40. [Crossref] [PubMed]
- Soria JC, Ohe Y, Vansteenkiste J, et al. Osimertinib in Untreated EGFR -Mutated Advanced Non-Small-Cell Lung Cancer. N Engl J Med 2018;378:113-25. [Crossref] [PubMed]
- Nagano T, Tachihara M, Nishimura Y. Mechanism of Resistance to Epidermal Growth Factor Receptor-Tyrosine Kinase Inhibitors and a Potential Treatment Strategy. Cells 2018;7:212. [Crossref] [PubMed]
- Dong J, Li B, Lin D, et al. Advances in Targeted Therapy and Immunotherapy for Non-small Cell Lung Cancer Based on Accurate Molecular Typing. Front Pharmacol 2019;10:230. [Crossref] [PubMed]
- Arulananda S, Do H, Musafer A, et al. Combination Osimertinib and Gefitinib in C797S and T790M EGFR-Mutated Non-Small Cell Lung Cancer. J Thorac Oncol 2017;12:1728-32. [Crossref] [PubMed]
- Schmid S, Li JJ, Leighl NB. Mechanisms of osimertinib resistance and emerging treatment options. Lung Cancer 2020;147:123-9. [Crossref] [PubMed]
- Bruno R, Proietti A, Alì G, et al. Squamous cell transformation and EGFR T790M mutation as acquired resistance mechanisms in a patient with lung adenocarcinoma treated with a tyrosine kinase inhibitor: A case report. Oncol Lett 2017;14:5947-51. [Crossref] [PubMed]
- Russo A, Franchina T, Ricciardi G, et al. Heterogeneous Responses to Epidermal Growth Factor Receptor (EGFR) Tyrosine Kinase Inhibitors (TKIs) in Patients with Uncommon EGFR Mutations: New Insights and Future Perspectives in this Complex Clinical Scenario. Int J Mol Sci 2019;20:1431. [Crossref] [PubMed]
- FDA Broadens Afatinib Indication to Previously Untreated, Metastatic NSCLC With Other Non-Resistant EGFR Mutations. FDA; 2018.
- Naidoo J, Sima CS, Rodriguez K, et al. Epidermal growth factor receptor exon 20 insertions in advanced lung adenocarcinomas: Clinical outcomes and response to erlotinib: EGFR Exon 20 Insertions. Cancer 2015;121:3212-20. [Crossref] [PubMed]
- Vyse S, Huang PH. Targeting EGFR exon 20 insertion mutations in non-small cell lung cancer. Signal Transduct Target Ther 2019;4:5. [Crossref] [PubMed]
- Robichaux JP, Elamin YY, Tan Z, et al. Mechanisms and clinical activity of an EGFR and HER2 exon 20-selective kinase inhibitor in non-small cell lung cancer. Nat Med 2018;24:638-46. [Crossref] [PubMed]
- Zaman A, Wu W, Bivona TG. Targeting Oncogenic BRAF: Past, Present, and Future. Cancers (Basel) 2019;11:1197. [Crossref] [PubMed]
- Dankner M, Rose AAN, Rajkumar S, et al. Classifying BRAF alterations in cancer: new rational therapeutic strategies for actionable mutations. Oncogene 2018;37:3183-99. [Crossref] [PubMed]
- Tissot C, Couraud S, Tanguy R, et al. Clinical characteristics and outcome of patients with lung cancer harboring BRAF mutations. Lung Cancer 2016;91:23-8. [Crossref] [PubMed]
- Planchard D, Smit EF, Groen HJM, et al. Dabrafenib plus trametinib in patients with previously untreated BRAFV600E-mutant metastatic non-small-cell lung cancer: an open-label, phase 2 trial. Lancet Oncol 2017;18:1307-16. [Crossref] [PubMed]
- Facchinetti F, Lacroix L, Mezquita L, et al. Molecular mechanisms of resistance to BRAF and MEK inhibitors in BRAFV600E non-small cell lung cancer. Eur J Cancer 2020;132:211-23. [Crossref] [PubMed]
- Drilon A, Jenkins C, Iyer S, et al. ROS1-dependent cancers - biology, diagnostics and therapeutics. Nat Rev Clin Oncol 2021;18:35-55. [Crossref] [PubMed]
- Mhanna L, Guibert N, Milia J, et al. When to Consider Immune Checkpoint Inhibitors in Oncogene-Driven Non-Small Cell Lung Cancer? Curr Treat Options Oncol 2019;20:60. [Crossref] [PubMed]
- Liang H, Wang M. MET Oncogene in Non-Small Cell Lung Cancer: Mechanism of MET Dysregulation and Agents Targeting the HGF/c-Met Axis. Onco Targets Ther 2020;13:2491-510. [Crossref] [PubMed]
- Kawakami H, Okamoto I, Okamoto W, et al. Targeting MET Amplification as a New Oncogenic Driver. Cancers (Basel) 2014;6:1540-52. [Crossref] [PubMed]
- Seo JS, Ju YS, Lee WC, et al. The transcriptional landscape and mutational profile of lung adenocarcinoma. Genome Res 2012;22:2109-19. [Crossref] [PubMed]
- Ma PC, Jagadeeswaran R, Jagadeesh S, et al. Functional Expression and Mutations of c-Met and Its Therapeutic Inhibition with SU11274 and Small Interfering RNA in Non-Small Cell Lung Cancer. Cancer Res 2005;65:1479-88. [Crossref] [PubMed]
- Champagnac A, Bringuier PP, Barritault M, et al. Frequency of MET exon 14 skipping mutations in non-small cell lung cancer according to technical approach in routine diagnosis: results from a real-life cohort of 2,369 patients. J Thorac Dis 2020;12:2172-8. [Crossref] [PubMed]
- Bylicki O, Paleiron N, Assié JB, et al. Targeting the MET-Signaling Pathway in Non-Small-Cell Lung Cancer: Evidence to Date. Onco Targets Ther 2020;13:5691-706. [Crossref] [PubMed]
- Salgia R, Sattler M, Scheele J, et al. The promise of selective MET inhibitors in non-small cell lung cancer with MET exon 14 skipping. Cancer Treat Rev 2020;87:102022. [Crossref] [PubMed]
- Moosavi F, Giovannetti E, Saso L, et al. HGF/MET pathway aberrations as diagnostic, prognostic, and predictive biomarkers in human cancers. Crit Rev Clin Lab Sci 2019;56:533-66. [Crossref] [PubMed]
- Merck KGaA. Merck Announces FDA Breakthrough Therapy Designation for Investigational Therapy Tepotinib in Patients with Metastatic NSCLC with METex14 Skipping Alterations. Available online: https://www.merckgroup.com/en/news/ tepotinib-breakthrough-therapy-designation-11-09-2019.html
- Li BT, Ross DS, Aisner DL, et al. HER2 Amplification and HER2 Mutation Are Distinct Molecular Targets in Lung Cancers. J Thorac Oncol 2016;11:414-9. [Crossref] [PubMed]
- Takezawa K, Pirazzoli V, Arcila ME, et al. HER2 Amplification: A Potential Mechanism of Acquired Resistance to EGFR Inhibition in EGFR -Mutant Lung Cancers That Lack the Second-Site EGFR T790M Mutation. Cancer Discov 2012;2:922-33. [Crossref] [PubMed]
- Guo Y, Cao R, Zhang X, et al. Recent Progress in Rare Oncogenic Drivers and Targeted Therapy For Non-Small Cell Lung Cancer. Onco Targets Ther 2019;12:10343-60. [Crossref] [PubMed]
- Koga T, Kobayashi Y, Tomizawa K, et al. Activity of a novel HER2 inhibitor, poziotinib, for HER2 exon 20 mutations in lung cancer and mechanism of acquired resistance: An in vitro study. Lung Cancer 2018;126:72-9. [Crossref] [PubMed]
- Chu QS. Targeting non-small cell lung cancer: driver mutation beyond epidermal growth factor mutation and anaplastic lymphoma kinase fusion. Ther Adv Med Oncol 2020;12:1758835919895756. [Crossref] [PubMed]
- Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 2007;448:561-6. [Crossref] [PubMed]
- Shaw AT, Yeap BY, Mino-Kenudson M, et al. Clinical Features and Outcome of Patients With Non-Small-Cell Lung Cancer Who Harbor EML4-ALK. J Clin Oncol 2009;27:4247-53. [Crossref] [PubMed]
- Thunnissen E, Bubendorf L, Dietel M, et al. EML4-ALK testing in non-small cell carcinomas of the lung: a review with recommendations. Virchows Archiv 2012;461:245-57. [Crossref] [PubMed]
- Yoshida T, Oya Y, Tanaka K, et al. Differential Crizotinib Response Duration Among ALK Fusion Variants in ALK -Positive Non-Small-Cell Lung Cancer. J Clin Oncol 2016;34:3383-9. [Crossref] [PubMed]
- Chen Y, Zhang X, Jiang Q, et al. Lung adenocarcinoma with a novel SRBD1-ALK Fusion responding to crizotinib. Lung Cancer 2020;146:370-2. [Crossref] [PubMed]
- Kwak EL, Bang YJ, Camidge DR, et al. Anaplastic Lymphoma Kinase Inhibition in Non-Small-Cell Lung Cancer. N Engl J Med 2010;363:1693-703. [Crossref] [PubMed]
- Shaw AT, Ou SHI, Bang YJ, et al. Crizotinib in ROS1 -Rearranged Non-Small-Cell Lung Cancer. N Engl J Med 2014;371:1963-71. [Crossref] [PubMed]
- Cortinovis D, Canova S, Abbate MI, et al. Challenges in ALK inhibition of ALK-positive non-small-cell lung cancer: from ALK positivity detection to treatment strategies after relapse. Future Oncol 2018;14:2303-17. [Crossref] [PubMed]
- Shaw AT, Friboulet L, Leshchiner I, et al. Resensitization to Crizotinib by the Lorlatinib ALK Resistance Mutation L1198F. N Engl J Med 2016;374:54-61. [Crossref] [PubMed]
- Landi L, Chiari R, Tiseo M, et al. Crizotinib in MET-Deregulated or ROS1-Rearranged Pretreated Non-Small Cell Lung Cancer (METROS): A Phase II, Prospective, Multicenter, Two-Arms Trial. Clin Cancer Res 2019;25:7312-9. [Crossref] [PubMed]
- Bergethon K, Shaw AT, Ignatius Ou SH, et al. ROS1 Rearrangements Define a Unique Molecular Class of Lung Cancers. J Clin Oncol 2012;30:863-70. [Crossref] [PubMed]
- Lim SM, Kim HR, Lee JS, et al. Open-Label, Multicenter, Phase II Study of Ceritinib in Patients With Non-Small-Cell Lung Cancer Harboring ROS1 Rearrangement. J Clin Oncol 2017;35:2613-8. [Crossref] [PubMed]
- Solomon BJ, Martini JF, Ou SH, et al. Efficacy of lorlatinib in patients (pts) with ROS1-positive advanced non-small cell lung cancer (NSCLC) and ROS1 kinase domain mutations. Ann Oncol 2018;29:viii495.
- Drilon A, Siena S, Dziadziuszko R, et al. Entrectinib in ROS1 fusion-positive non-small-cell lung cancer: integrated analysis of three phase 1-2 trials. Lancet Oncol 2020;21:261-70. [Crossref] [PubMed]
- Cho BC, Drilon AE, Doebele RC, et al. Safety and preliminary clinical activity of repotrectinib in patients with advanced ROS1 fusion-positive non-small cell lung cancer (TRIDENT-1 study). J Clin Oncol 2019;37:9011. [Crossref]
- Ju YS, Lee WC, Shin JY, et al. A transforming KIF5B and RET gene fusion in lung adenocarcinoma revealed from whole-genome and transcriptome sequencing. Genome Res 2012;22:436-45. [Crossref] [PubMed]
- Ferrara R, Auger N, Auclin E, et al. Clinical and Translational Implications of RET Rearrangements in Non-Small Cell Lung Cancer. J Thorac Oncol 2018;13:27-45. [Crossref] [PubMed]
- Michels S, Scheel AH, Scheffler M, et al. Clinicopathological Characteristics of RET Rearranged Lung Cancer in European Patients. J Thorac Oncol 2016;11:122-7. [Crossref] [PubMed]
- Drilon A, Oxnard G, Wirth L, et al. PL02.08 Registrational Results of LIBRETTO-001: A Phase 1/2 Trial of LOXO-292 in Patients with RET Fusion-Positive Lung Cancers. J Thorac Oncol 2019;14:S6-S7. [Crossref]
- Drilon A, Rekhtman N, Arcila M, et al. Cabozantinib in patients with advanced RET -rearranged non-small-cell lung cancer: an open-label, single-centre, phase 2, single-arm trial. Lancet Oncol 2016;17:1653-60. [Crossref] [PubMed]
- Amatu A, Sartore-Bianchi A, Bencardino K, et al. Tropomyosin receptor kinase (TRK) biology and the role of NTRK gene fusions in cancer. Ann Oncol 2019;30:viii5-viii15.
- Cocco E, Scaltriti M, Drilon A. NTRK fusion-positive cancers and TRK inhibitor therapy. Nat Rev Clin Oncol 2018;15:731-47. [Crossref] [PubMed]
- Solomon JP, Benayed R, Hechtman JF, et al. Identifying patients with NTRK fusion cancer. Ann Oncol 2019;30:viii16-viii22.
- Farago AF, Kummar S, Moreno V, et al. B04 activity of Larotrectinib in tropomyosin receptor kinase fusion lung Cancer. J Thorac Oncol 2020;15:25. [Crossref]
- Doebele RC, Drilon A, Paz-Ares L, et al. Entrectinib in patients with advanced or metastatic NTRK fusion-positive solid tumours: integrated analysis of three phase 1-2 trials. Lancet Oncol 2020;21:271-82. [Crossref] [PubMed]
- Lau J, Cheung J, Navarro A, et al. Tumour and host cell PD-L1 is required to mediate suppression of anti-tumour immunity in mice. Nat Commun 2017;8:14572. [Crossref] [PubMed]
- Yuasa T, Masuda H, Yamamoto S, et al. Biomarkers to predict prognosis and response to checkpoint inhibitors. Int J Clin Oncol 2017;22:629-34. [Crossref] [PubMed]
- Lantuejoul S, Sound-Tsao M, Cooper WA, et al. PD-L1 Testing for Lung Cancer in 2019: Perspective From the IASLC Pathology Committee. J Thorac Oncol 2020;15:499-519. [Crossref] [PubMed]
- Garon EB, Rizvi NA, Hui R, et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med 2015;372:2018-28. [Crossref] [PubMed]
- Reck M, Rodríguez-Abreu D, Robinson AG, et al. Updated Analysis of KEYNOTE-024: Pembrolizumab Versus Platinum-Based Chemotherapy for Advanced Non-Small-Cell Lung Cancer With PD-L1 Tumor Proportion Score of 50% or Greater. J Clin Oncol 2019;37:537-46. [Crossref] [PubMed]
- Mok TSK, Wu YL, Kudaba I, et al. Pembrolizumab versus chemotherapy for previously untreated, PD-L1-expressing, locally advanced or metastatic non-small-cell lung cancer (KEYNOTE-042): a randomised, open-label, controlled, phase 3 trial. Lancet 2019;393:1819-30. [Crossref] [PubMed]
- Reck M, Rodríguez-Abreu D, Robinson AG, et al. Pembrolizumab versus Chemotherapy for PD-L1-Positive Non-Small-Cell Lung Cancer. N Engl J Med 2016;375:1823-33. [Crossref] [PubMed]
- Gadgeel S, Rodríguez-Abreu D, Speranza G, et al. Updated Analysis From KEYNOTE-189: Pembrolizumab or Placebo Plus Pemetrexed and Platinum for Previously Untreated Metastatic Nonsquamous Non-Small-Cell Lung Cancer. J Clin Oncol 2020;38:1505-17. [Crossref] [PubMed]
- Paz-Ares L, Luft A, Vicente D, et al. Pembrolizumab plus Chemotherapy for Squamous Non-Small-Cell Lung Cancer. N Engl J Med 2018;379:2040-51. [Crossref] [PubMed]
- Brahmer J, Reckamp KL, Baas P, et al. Nivolumab versus Docetaxel in Advanced Squamous-Cell Non-Small-Cell Lung Cancer. N Engl J Med 2015;373:123-35. [Crossref] [PubMed]
- Borghaei H, Paz-Ares L, Horn L, et al. Nivolumab versus Docetaxel in Advanced Nonsquamous Non-Small-Cell Lung Cancer. N Engl J Med 2015;373:1627-39. [Crossref] [PubMed]
- Fehrenbacher L, Spira A, Ballinger M, et al. Atezolizumab versus docetaxel for patients with previously treated non-small-cell lung cancer (POPLAR): a multicentre, open-label, phase 2 randomised controlled trial. Lancet 2016;387:1837-46. [Crossref] [PubMed]
- Rittmeyer A, Barlesi F, Waterkamp D, et al. Atezolizumab versus docetaxel in patients with previously treated non-small-cell lung cancer (OAK): a phase 3, open-label, multicentre randomised controlled trial. Lancet 2017;389:255-65. [Crossref] [PubMed]
- Rizvi NA, Hellmann MD, Snyder A, et al. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015;348:124-8. [Crossref] [PubMed]
- Hu H, She L, Liao M, et al. Cost-Effectiveness Analysis of Nivolumab Plus Ipilimumab vs. Chemotherapy as First-Line Therapy in Advanced Non-Small Cell Lung Cancer. Front Oncol 2020;10:1649. [Crossref] [PubMed]
- Ayers M, Lunceford J, Nebozhyn M, et al. IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest 2017;127:2930-40. [Crossref] [PubMed]
- Kerr KM, López-Ríos F. Precision medicine in NSCLC and pathology: how does ALK fit in the pathway? Ann Oncol 2016;27:iii16-iii24. [Crossref] [PubMed]
- Mosele F, Remon J, Mateo J, et al. Recommendations for the use of next-generation sequencing (NGS) for patients with metastatic cancers: a report from the ESMO Precision Medicine Working Group. Ann Oncol 2020;31:1491-505. [Crossref] [PubMed]
- Lim ZF, Ma PC. Emerging insights of tumor heterogeneity and drug resistance mechanisms in lung cancer targeted therapy. J Hematol Oncol 2019;12:134. [Crossref] [PubMed]
- Guibert N, Pradines A, Favre G, et al. Current and future applications of liquid biopsy in nonsmall cell lung cancer from early to advanced stages. Eur Respir Rev 2020;29:190052. [Crossref] [PubMed]
- Tachon G, Cortes U, Richard S, et al. Targeted RNA‐sequencing assays: a step forward compared to FISH and IHC techniques? Cancer Med 2019;8:7556-66. [Crossref] [PubMed]
- Solomon JP, Linkov I, Rosado A, et al. NTRK fusion detection across multiple assays and 33,997 cases: diagnostic implications and pitfalls. Mod Pathol 2020;33:38-46. [Crossref] [PubMed]
- Armstrong F, Duplantier MM, Trempat P, et al. Differential effects of X-ALK fusion proteins on proliferation, transformation, and invasion properties of NIH3T3 cells. Oncogene 2004;23:6071-82. [Crossref] [PubMed]
- Bruno R, Fontanini G. Next Generation Sequencing for Gene Fusion Analysis in Lung Cancer: A Literature Review. Diagnostics 2020;10:521. [Crossref] [PubMed]
- Rogers TM, Arnau GM, Ryland GL, et al. Multiplexed transcriptome analysis to detect ALK, ROS1 and RET rearrangements in lung cancer. Sci Rep 2017;7:42259. [Crossref] [PubMed]
- Alì G, Bruno R, Savino M, et al. Analysis of Fusion Genes by NanoString System: A Role in Lung Cytology? Arch Pathol Lab Med 2018;142:480-9. [Crossref] [PubMed]
- Allegretti M, Fabi A, Buglioni S, et al. Tearing down the walls: FDA approves next generation sequencing (NGS) assays for actionable cancer genomic aberrations. J Exp Clin Cancer Res 2018;37:47. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin 2017;67:7-30. [Crossref] [PubMed]
- Domagala-Kulawik J, Trojnar A. Lung cancer in women in 21th century. J Thorac Dis 2020;12:4398-410. [Crossref] [PubMed]
- MacRosty CR, Rivera MP. Lung Cancer in Women. Clin Chest Med 2020;41:53-65. [Crossref] [PubMed]
- O’Keeffe LM, Taylor G, Huxley RR, et al. Smoking as a risk factor for lung cancer in women and men: a systematic review and meta-analysis. BMJ Open 2018;8:e021611. [Crossref] [PubMed]
- Tang DL. Associations between both genetic and environmental biomarkers and lung cancer: evidence of a greater risk of lung cancer in women smokers. Carcinogenesis 1998;19:1949-53. [Crossref] [PubMed]
- Toyooka S, Tsuda T, Gazdar AF. TheTP53 gene, tobacco exposure, and lung cancer. Hum Mutat 2003;21:229-39. [Crossref] [PubMed]
- North CM, Christiani DC. Women and Lung Cancer: What is New? Semin Thorac Cardiovasc Surg 2013;25:87-94. [Crossref] [PubMed]
- Kligerman S, White C. Epidemiology of Lung Cancer in Women: Risk Factors, Survival, and Screening. AJR Am J Roentgenol 2011;196:287-95. [Crossref] [PubMed]
- Testa U, Castelli G, Pelosi E. Lung Cancers: Molecular Characterization, Clonal Heterogeneity and Evolution, and Cancer Stem Cells. Cancers (Basel) 2018;10:248. [Crossref] [PubMed]
- Kadota K, Yeh YC, D’Angelo SP, et al. Associations Between Mutations and Histologic Patterns of Mucin in Lung Adenocarcinoma: Invasive Mucinous Pattern and Extracellular Mucin Are Associated With KRAS Mutation. Am J Surg Pathol 2014;38:1118-27. [Crossref] [PubMed]
- Liu L, Shao X, Gao W, et al. The Role of Human Epidermal Growth Factor Receptor 2 as a Prognostic Factor in Lung Cancer: A Meta-Analysis of Published Data. J Thorac Oncol 2010;5:1922-32. [Crossref] [PubMed]
- Dogan S, Shen R, Ang DC, et al. Molecular Epidemiology of EGFR and KRAS Mutations in 3,026 Lung Adenocarcinomas: Higher Susceptibility of Women to Smoking-Related KRAS-Mutant Cancers. Clin Cancer Res 2012;18:6169-77. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511:543-50. [Crossref] [PubMed]
- Jaeger N, Czepielewski RS, Bagatini M, et al. Neuropeptide gastrin-releasing peptide induces PI3K/reactive oxygen species-dependent migration in lung adenocarcinoma cells. Tumour Biol 2017;39:1010428317694321. [Crossref] [PubMed]
- Li Y, He CL, Li WX, et al. Transcriptome analysis reveals gender-specific differences in overall metabolic response of male and female patients in lung adenocarcinoma. PLoS One 2020;15:e0230796. [Crossref] [PubMed]
- Özdemir BC, Dotto GP. Sex Hormones and Anticancer Immunity. Clin Cancer Res 2019;25:4603-10. [Crossref] [PubMed]
- Rodriguez-Lara V, Peña-Mirabal E, Baez-Saldaña R, et al. Estrogen Receptor Beta and CXCR4/CXCL12 Expression: Differences by Sex and Hormonal Status in Lung Adenocarcinoma. Arch Med Res 2014;45:158-69. [Crossref] [PubMed]
- Massaro D, Massaro GD. Estrogen receptor regulation of pulmonary alveolar dimensions: alveolar sexual dimorphism in mice. A Am J Physiol Lung Cell Mol Physiol 2006;290:L866-L870. [Crossref] [PubMed]
- Deng F, Li M, Shan WL, et al. Correlation between epidermal growth factor receptor mutations and the expression of estrogen receptor-β in advanced non-small cell lung cancer. Oncol Lett 2017;13:2359-65. [Crossref] [PubMed]
- Cheng TD, Darke AK, Redman MW, et al. Smoking, Sex, and Non-Small Cell Lung Cancer: Steroid Hormone Receptors in Tumor Tissue (S0424). J Natl Cancer Inst 2018;110:734-42. [Crossref] [PubMed]
- Dubois C, Rocks N, Blacher S, et al. Lymph/angiogenesis contributes to sex differences in lung cancer through oestrogen receptor alpha signalling. Endocr Relat Cancer 2019;26:201-16. [Crossref] [PubMed]
- Liao J, McCauley LK. Skeletal metastasis: Established and emerging roles of parathyroid hormone related protein (PTHrP). Cancer Metastasis Rev 2006;25:559-71. [Crossref] [PubMed]
- Hastings RH. Sex-Specific Survival Advantage with Parathyroid Hormone-Related Protein in Non-Small Cell Lung Carcinoma Patients. Clin Cancer Res 2006;12:499-506. [Crossref] [PubMed]
- Duma N, Abdel‐Ghani A, Yadav S, et al. Sex Differences in Tolerability to Anti-Programmed Cell Death Protein 1 Therapy in Patients with Metastatic Melanoma and Non-Small Cell Lung Cancer: Are We All Equal? Oncologist 2019;24:e1148-e1155. [Crossref] [PubMed]
- Caetano MS, Hassane M, Van HT, et al. Sex specific function of epithelial STAT3 signaling in pathogenesis of K-ras mutant lung cancer. Nat Commun 2018;9:4589. [Crossref] [PubMed]
- Wang L, Cao Y, Ren M, et al. Sex Differences in Hazard Ratio During Drug Treatment of Non-small-cell Lung Cancer in Major Clinical Trials: A Focused Data Review and Meta-analysis. Clin Ther 2017;39:34-54. [Crossref] [PubMed]
- Yuan Y, Liu L, Chen H, et al. Comprehensive Characterization of Molecular Differences in Cancer between Male and Female Patients. Cancer Cell 2016;29:711-22. [Crossref] [PubMed]
- Janku F, Kaseb AO, Tsimberidou AM, et al. Identification of novel therapeutic targets in the PI3K/AKT/mTOR pathway in hepatocellular carcinoma using targeted next generation sequencing. Oncotarget 2014;5:3012-22. [Crossref] [PubMed]
- Shackelford DB, Abt E, Gerken L, et al. LKB1 Inactivation Dictates Therapeutic Response of Non-Small Cell Lung Cancer to the Metabolism Drug Phenformin. Cancer Cell 2013;23:143-58. [Crossref] [PubMed]
- Chen Z, Zhang J, Huang K, et al. Comparison of clinicopathologic characteristics between patients with EGFR exon 19 deletion and EGFR L858R mutation in lung cancer. Int J Clin Exp Pathol 2018;11:4644-9. [PubMed]
- Koyama N, Watanabe Y, Iwai Y, et al. Distinct Benefit of Overall Survival between Patients with Non-Small-Cell Lung Cancer Harboring EGFR Exon 19 Deletion and Exon 21 L858R Substitution. Chemotherapy 2017;62:151-8. [Crossref] [PubMed]
- Zhang Y, Chen G, Chen X, et al. The comparison of EGFR-TKI failure modes and subsequent management between exon 19 deletion and exon 21 L858R mutation in advanced non-small-cell lung cancer. J Cancer 2017;8:1865-71. [Crossref] [PubMed]
- Spoerke JM, O’Brien C, Huw L, et al. Phosphoinositide 3-Kinase (PI3K) Pathway Alterations Are Associated with Histologic Subtypes and Are Predictive of Sensitivity to PI3K Inhibitors in Lung Cancer Preclinical Models. Clin Cancer Res 2012;18:6771-83. [Crossref] [PubMed]
- Chaft JE, Arcila ME, Paik PK, et al. Coexistence of PIK3CA and Other Oncogene Mutations in Lung Adenocarcinoma-Rationale for Comprehensive Mutation Profiling. Mol Cancer Ther 2012;11:485-91. [Crossref] [PubMed]
Cite this article as: Bruno R, Alì G, Poma AM, Fontanini G. Non-small cell lung cancer molecular characterization of advanced disease with focus on sex differences: a narrative review. Precis Cancer Med 2021;4:14.


