
Prognostic impact of targetable genetic variants in resected adenocarcinoma of the lung: a narrative review and model proposal for precise evaluation
Introduction
Targetable genetic variants have been discovered in non-small cell lung cancer (NSCLC), especially in adenocarcinoma. Newly developed drugs have more favorable results in adenocarcinoma treatment than the older ones. Targetable variants have become crucial for choosing treatment in advanced cases. In early-stage cases, surgery is the primary curative treatment, and the prognostic impact of targetable variants after resection remains controversial. Recently, ADAURA trial (NCT02511106), phase III, double-blind, randomized clinical trial estimating the utility of adjuvant osimertinib showed the promising results. Adjuvant osimertinib improved disease-free survival in resected stage IB, II, IIIA EGFR-mutant cases (1). Revealing prognostic implication of targetable variant and stratifying cases by the risk of recurrence or benefit by treatment will be more important. Herein, we review the literature concerning the prognostic impact of targetable genetic variants in resected lung cancer and propose a methodology for its precise evaluation. We present the following article in accordance with the Narrative Review reporting checklist (available at http://dx.doi.org/10.21037/pcm-20-47).
Source and selection criteria
A review of literature through PubMed was conducted using the keywords “non-small cell lung cancer” or “lung adenocarcinoma” with a gene name with targetable variants. Literature estimating resected NSCLC was reviewed, and relevant material was explored individually through cross-references. Studies estimating non-resected or advanced lung cancer only were excluded from the review. This study focused on the gene variants that were druggable targets, such as EGFR/BRAF mutations and ALK fusion. The impact of KRAS mutation was also searched as a frequent and promising target variant.
Summary and interpretation
The manuscripts chosen for review are shown in Table 1. All studies included surgically treated adenocarcinoma cases. The studies already included in the meta-analysis are fundamentally excluded from Table 1.
Table 1
| Analyzed gene | Author [year] | Number of analyzed cases (% of variant cases) | pStage (TNM edition) | Prognostic impact of genetic variant | Reference |
|---|---|---|---|---|---|
| EGFR | Zhang [2014]† | 3,337 (12.2–52.5) | I–IIIA (−) | No significance on OS and DFS | (2) |
| Takamochi [2017] | 741 (47.2) | I (IASLC, 7th) | longer OS | (3) | |
| Ito [2018] | 279 (47.0) | pN0, IA1–IIA (IASLC, 8th) | Shorter RFI | (4) | |
| He [2019]† | 4,872 (42.8) | I–IIIA (−) | No significance on DFS | (5) | |
| Ito [2020] | 664 (48.9) | ≤5 cm, pN0–1, IA1–IIIA (IASLC, 8th) | Shorter RFS | (6) | |
| Suda [2020] | 5,780 (41.7) | I–IV (−) | Better RFS and OS | (7) | |
| Deng [2020] | 1,249 (62.4) | I–III (unknown, 8th) | Worse RFS in radiological solid, Pap/Aci/IMA, or stage II–III | (8) | |
| KRAS | Meng [2013]‡ | 6,939 (4.5–36.4) | I–V (−) | Poorer prognostic status in adenocarcinoma, stage I, or stage I–IIIA | (9) |
| Ma [2020] | 1,181 (9.3) | I–III (unknown, 8th) | Poorer RFS and OS in part-solid or stage I | (10) | |
| ALK | Fukui [2012] | 720 (3.9) | IA–IV (unknown) | No significance in DFS and OS | (11) |
| Blackhall [2014] | 1,281 (2.2–6.2) | I–III (unknown) | Significantly better OS, no significance about RFS | (12) | |
| Gao [2017] | 534 (7.9) | I–III (unknown) | Shorter DFS, TSS, OS | (13) | |
| Shin [2018] | 309 (7.4) | IA (AJCC, 7th) | Lower DFS | (14) | |
| Liu [2019] | 81 (3.9) | I–IIIA (unknown, 8th) | No significance in DFS and OS, but higher risk of liver recurrence | (15) | |
| EGFR/KRAS | Ohba [2014] | 354 (41.1/6.4) | I (unknown) | Shorter DFS and OS in KRAS (+) | (16) |
| Izar [2014] | 312 (19.6/40.7) | I (AJCC, 7th) | Worse DFS and OS in KRAS (+) | (17) | |
| Zhang [2018]§ | 10,869 (33.6/15.5) | I–V (−) | Prolonged DFS and OS in EGFR (+), worse DFS and OS in KRAS (+) | (18) | |
| EGFR/KRAS/BRAF | Marchetti [2011] | 739 (11.6/27.5/4.9) | I–IV (unknown) | Shorter median DFS and OS in BRAF V600E (+) | (19) |
| Kadota [2016] | 482 (17.8/26.8/1.7) | I–II (AJCC, 7th) | Worse OS in KRAS (+), tendency of better OS in EGFR (+) | (20) | |
| Kneuertz [2020] | 324 (17.9/38.1/5.9) | IA–IIIB (AJCC, 8th) | Worse DFS and OS in KRAS (+) or BRAF (+) | (21) | |
| EGFR/KRAS/ALK/BRAF | Ohba [2016] | 256 (46.8/5.5/2.3/0.8) | I (unknown) | Worse DFS and OS in KRAS (+) | (22) |
| EGFR/KRAS/ALK | Chaft [2018]§ | 764 (33.4/62.8/3.8) | I–III (AJCC, 7th) | Worse RFS in ALK (+) than in EGFR (+) | (23) |
†, meta-analysis; ‡, meta-analysis including non-resected cases; §, including non-adenocarcinoma cases. Aci, acinar predominant adenocarcinoma; AJCC, American Joint Committee on Cancer; DFS, disease-free survival; IASLC, International Association for the Study of Lung Cancer; IMA, invasive mucinous adenocarcinoma; OS, overall survival; Pap, papillary predominant adenocarcinoma; RFI, recurrence-free interval; RFS, recurrence-free survival.
Prognostic impact of EGFR mutation
Three systematic meta-analyses concluded with conflicting results. Two analyses concluded that the EGFR mutation has no significant impact on the prognosis of adenocarcinoma (2,5), while the other suggested that the EGFR mutation is related to better prognosis (18). Systematic meta-analyses reviewed the literature thoroughly, and therefore inevitably included limitations. Several studies analyzed early and advanced stage cases as well as the non-adenocarcinoma cases while some considered adenocarcinoma to be of a single histological phenotype. The controversy about the impact of EGFR mutations has continued even after these meta-analyses. Two studies with large sample sizes suggested that the EGFR mutation is related to a worse prognosis (3,7). Another study that included non-adenocarcinoma cases indicated that the impact of EGFR mutation is limited in stage IB, with no impact on stage 0–IA (24). As a strong influencing factor on prognosis, the tumor-node-metastasis (TNM) staging system and pathological classification have been revised. The subtypes of adenocarcinoma were proposed in 2011 (25), therefore literature published before 2011 could not distinguish the histological subtypes of adenocarcinoma. Recently, our group and another group demonstrated EGFR mutation status as a poor prognostic factor in resected lung cancer while taking into consideration the pStage, histological subtype, and/or radiological features (4,6,8). EGFR mutations are more frequently harbored in cases involving lepidic lesions (4,6-8,20,26), which are histological features related to better prognosis. Therefore, it is rational that the studies that did not account for the recurrence risk by histological subtypes of adenocarcinoma concluded that EGFR mutation is a favorable prognostic factor. The majority of the EGFR-mutated cases in these studies could have been harbored in cases completely or almost free from relapse, such as adenocarcinoma in situ (AIS), minimally invasive adenocarcinoma (MIA), or lepidic predominant cases. Although quantitative estimation of histological components is highly recommended in resected cases (25), most studies only evaluated the pStage and not the histological subtypes of adenocarcinoma. Currently, there is no evidence regarding whether pStage or histological subtype is more important in invasive lung adenocarcinoma cases. More studies considering the pStage and the recurrence risk based on the histological subtypes of adenocarcinoma are warranted for precise estimation of the prognostic impact of genetic variants.
Prognostic impact of KRAS mutation
A systematic meta-analysis concluded that KRAS mutations are associated with worse overall survival (OS), especially in adenocarcinoma and early-stage cases (9). Another meta-analysis concluded similar results (18). Most studies published later also reported KRAS mutation status to be related to worse prognosis even when evaluated with other variants (16-18,20-22). KRAS mutation is likely to be harbored in cases involving a solid component (10,26,27). Solid predominant subtypes are high malignant phenotype (28,29), and the presence of solid component is also an unfavorable feature in prognosis even when it is not predominant (30,31). Therefore, poorer prognosis in KRAS mutant cases might reflect high malignant behavior stemming from the solid component. The high frequency of KRAS mutations in cases involving solid components might be akin to the tendency of wild-type KRAS in cases without solid components, such as AIS/MIA/lepidic predominant subtypes. To precisely determine whether the prognostic value of KRAS mutant cases comes from the mutation itself or the solid component, the histological subtypes should also be distinguished.
Prognostic impact of ALK fusion or BRAF mutation
ALK fusion is likely to be harbored in younger and/or more advanced cases (11-14,23) and histologically related to solid predominant or invasive mucinous adenocarcinoma (12,13). BRAF mutations are associated with adenocarcinoma with micropapillary components (19,32). The problem concerning ALK fusion or BRAF mutation is the low prevalence in general. Most studies estimating the impact of ALK fusion or BRAF mutation alone or with other variations reported worse prognostic tendency in ALK or BRAF variant cases (13,14,19,23). However, due to the low frequency of the ALK fusion or BRAF mutation, more validation studies with larger sample sizes are warranted. Although the analysis using propensity score matching (PSM) might be a useful methodology, especially in rare variants, two studies that used PSM did not indicate significant results in disease-free survival (DFS) or recurrence-free survival (RFS) for ALK fusion (12,15).
Estimation of several genetic variants
Most studies evaluated a single gene and discussed the prognostic impact (Table 1). In general, studies evaluating several genetic variants simultaneously are more reliable than those that study single genes. A comparison of EGFR-mutant and EGFR wild type in mono-gene analysis is insufficient to estimate the impact of EGFR mutation. The ‘wild type’ may include KRAS-mutant, BRAF-mutant, ALK-rearrangement, and other targetable variants. Labeling cases not involving EGFR mutation as ‘Wild type’ is risky, as the effects of another variant may be overlooked.
EGFR mutations are one of the most frequent targetable variants. KRAS mutations have not been clinically targetable yet. However, KRAS mutation includes a promising druggable variant (33), and most studies concluded that KRAS mutation leads to worse prognostic status. Therefore, it might be better to evaluate the prognostic impact of genetic variants in multi-gene analysis, including at least EGFR and KRAS mutations.
Discussion
As a prognostic reflector, the TNM staging system is the gold standard. It covers all phases of tumorigenesis, from in situ to multiple distant metastases. Histological classification is another standard for predicting prognosis, especially in resected cases. As staging and pathological features can vary according to the timing of diagnosis, they can reflect prognosis in detail. On the other hand, the prognostic impact of genetic variants remains controversial. Compared to the TNM staging or pathological classification, evaluating genetic status is more challenging due to various factors, such as cost, wide variety of variants, different sensitivity of methodologies, the necessity for a larger sample volume. In individual cases, the genetic status is fundamentally unchanged by the timing of diagnosis and is labeled simply as positive or negative. It is the strength and weakness of genetic variants as prognostic factors; genetic status is objective and quantitative, but too simple to reflect the change in the malignant phase. Nevertheless, genetic variants have the advantage of being useful in the choice of treatment for adenocarcinoma in case of recurrence. It must be noted that the distribution of genetic variants can differ according to the stage and pathological features. Because staging and pathological diagnosis are rigid standards and reflect the disease prognosis more sensitively, the impact of genetic variants should be considered after estimating staging and pathological features. Even for the KRAS mutation, which is the most promising prognostic marker in genetic variants, one of the latest studies suggested that considering stage and clinicopathological features is crucial for its use as a prognostic predictor (10).
We propose a distribution model of EGFR and KRAS mutations according to the stage and pathological status (Figure 1). Based on this, we propose an ideal methodology for estimating the prognostic impact of genetic variants by considering histological variants. Briefly, the stage and histological subtype should be matched, and several variants should be estimated (Figure 2A). However, it is not easy to perform comprehensive genetic analysis and collect a balanced number of cases. In case a mono-gene comparison is performed, we must exclude the improper setting (Figure 2B,C) as much as possible.
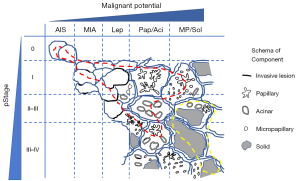
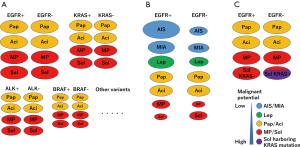
Apart from the abovementioned problem, attention must be paid to several other issues. To this end, the frequency of genetic variants differs among races or regions. It must be noted that even major variants, such as EGFR or KRAS mutations can be low frequent variants, such as BRAF mutation or ALK rearrangement and the prognostic impact can be underestimated in different races or regions.
OS is prolonged by target therapy after recurrence (34,35). To estimate the prognostic impact of genetic variants in a natural course, OS is not always a proper endpoint. We also have to consider race and treatment course in estimating the genetic prognostic value.
Conclusions
The staging system and pathological classification sensitively reflect the prognosis. The distribution of genetic status varies by stage, histological features, and race, especially in adenocarcinoma. Although genetic variants are promising prognostic predictors, their use is currently limited and can be misinterpreted unless they are considered along with stage and histology. Further discussion should be conducted by considering stage and histological features with multi-gene analysis studies.
Acknowledgments
The authors appreciate the Editor for providing the wonderful opportunity to submit this invited review article. The authors thank Editage (www.editage.jp) for English language editing.
Funding: None.
Footnote
Reporting Checklist: The authors have completed the Narrative Review reporting checklist. Available at http://dx.doi.org/10.21037/pcm-20-47
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/pcm-20-47). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Herbst RS, Tsuboi M, John T, et al. Osimertinib as adjuvant therapy in patients (pts) with stage IB–IIIA EGFR mutation positive (EGFRm) NSCLC after complete tumor resection: ADAURA. J Clin Oncol 2020;38:LBA5. [Crossref]
- Zhang Z, Wang T, Zhang J, et al. Prognostic value of epidermal growth factor receptor mutations in resected non-small cell lung cancer: a systematic review with meta-analysis. PLoS One 2014;9:e106053. [Crossref] [PubMed]
- Takamochi K, Oh S, Matsunaga T, et al. Prognostic impacts of EGFR mutation status and subtype in patients with surgically resected lung adenocarcinoma. J Thorac Cardiovasc Surg 2017;154:1768-74.e1. [Crossref] [PubMed]
- Ito M, Miyata Y, Kushitani K, et al. Increased risk of recurrence in resected EGFR-positive pN0M0 invasive lung adenocarcinoma. Thorac Cancer 2018;9:1594-602. [Crossref] [PubMed]
- He Q, Xin P, Zhang M, et al. The impact of epidermal growth factor receptor mutations on the prognosis of resected non-small cell lung cancer: A meta-analysis of literatures. Transl Lung Cancer Res 2019;8:124-34. [Crossref] [PubMed]
- Ito M, Miyata Y, Tsutani Y, et al. Positive EGFR mutation status is a risk of recurrence in pN0-1 lung adenocarcinoma when combined with pathological stage and histological subtype: A retrospective multi-center analysis. Lung Cancer 2020;141:107-13. [Crossref] [PubMed]
- Suda K, Mitsudomi T, Shintani Y, et al. Clinical impacts of EGFR mutation status: analysis of 5,780 surgically resected lung cancer cases. Ann Thorac Surg 2020; [Epub ahead of print]. [Crossref] [PubMed]
- Deng C, Zhang Y, Ma Z, et al. Prognostic value of epidermal growth factor receptor gene mutation in resected lung adenocarcinoma. J Thorac Cardiovasc Surg 2020; [Epub ahead of print]. [Crossref] [PubMed]
- Meng D, Yuan M, Li X, et al. Prognostic value of K-RAS mutations in patients with non-small cell lung cancer: A systematic review with meta-analysis. Lung Cancer 2013;81:1-10. [Crossref] [PubMed]
- Ma Z, Zhang Y, Deng C, et al. The prognostic value of Kirsten rat sarcoma viral oncogene homolog mutations in resected lung adenocarcinoma differs according to clinical features. J Thorac Cardiovasc Surg 2020; [Epub ahead of print]. [Crossref] [PubMed]
- Fukui T, Yatabe Y, Kobayashi Y, et al. Clinicoradiologic characteristics of patients with lung adenocarcinoma harboring EML4-ALK fusion oncogene. Lung Cancer 2012;77:319-25. [Crossref] [PubMed]
- Blackhall FH, Peters S, Bubendorf L, et al. Prevalence and clinical outcomes for patients with ALK-positive resected stage I to III adenocarcinoma: results from the European Thoracic Oncology Platform Lungscape Project. J Clin Oncol 2014;32:2780-7. [Crossref] [PubMed]
- Gao Q, Li P, Jiang X, et al. Worse disease-free, tumor-specific, and overall survival in surgically-resected lung adenocarcinoma patients with ALK rearrangement. Oncotarget 2017;8:86066-81. [Crossref] [PubMed]
- Shin SH, Lee H, Jeong BH, et al. Anaplastic lymphoma kinase rearrangement in surgically resected stage IA lung adenocarcinoma. J Thorac Dis 2018;10:3460-7. [Crossref] [PubMed]
- Liu Y, Ye X, Yu Y, et al. Prognostic significance of anaplastic lymphoma kinase rearrangement in patients with completely resected lung adenocarcinoma. J Thorac Dis 2019;11:4258-70. [Crossref] [PubMed]
- Ohba T, Toyokawa G, Kometani T, et al. Mutations of the EGFR and K-ras genes in resected stage I lung adenocarcinoma and their clinical significance. Surg Today 2014;44:478-86. [Crossref] [PubMed]
- Izar B, Zhou H, Heist RS, et al. The prognostic impact of KRAS, its codon and amino acid specific mutations, on survival in resected stage I lung adenocarcinoma. J Thorac Oncol 2014;9:1363-9. [Crossref] [PubMed]
- Zhang SM, Zhu QG, Ding XX, et al. Prognostic value of EGFR and KRAS in resected non-small cell lung cancer: a systematic review and meta-analysis. Cancer Manag Res 2018;10:3393-404. [Crossref] [PubMed]
- Marchetti A, Felicioni L, Malatesta S, et al. Clinical features and outcome of patients with non-small-cell lung cancer harboring BRAF mutations. J Clin Oncol 2011;29:3574-9. [Crossref] [PubMed]
- Kadota K, Sima CS, Arcila ME, et al. KRAS mutation is a significant prognostic factor in early-stage lung adenocarcinoma. Am J Surg Pathol 2016;40:1579-90. [Crossref] [PubMed]
- Kneuertz PJ, Carbone DP, D'Souza DM, et al. Prognostic value and therapeutic implications of expanded molecular testing for resected early stage lung adenocarcinoma. Lung Cancer 2020;143:60-6. [Crossref] [PubMed]
- Ohba T, Toyokawa G, Osoegawa A, et al. Mutations of the EGFR, K-ras, EML4-ALK, and BRAF genes in resected pathological stage I lung adenocarcinoma. Surg Today 2016;46:1091-8. [Crossref] [PubMed]
- Chaft JE, Dagogo-Jack I, Santini FC, et al. Clinical outcomes of patients with resected, early-stage ALK-positive lung cancer. Lung Cancer 2018;122:67-71. [Crossref] [PubMed]
- Isaka T, Ito H, Nakayama H, et al. Effect of epidermal growth factor receptor mutation on early-stage non-small cell lung cancer according to the 8th TNM classification. Lung Cancer 2020;145:111-8. [Crossref] [PubMed]
- Travis WD, Brambilla E, Noguchi M, et al. International association for the study of lung cancer/American Thoracic Society/European Respiratory Society international multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol 2011;6:244-85. [Crossref] [PubMed]
- Yoshizawa A, Sumiyoshi S, Sonobe M, et al. Validation of the IASLC/ATS/ERS lung adenocarcinoma classification for prognosis and association with EGFR and KRAS gene mutations: Analysis of 440 Japanese patients. J Thorac Oncol 2013;8:52-61. [Crossref] [PubMed]
- Rekhtman N, Ang DC, Riely GJ, et al. KRAS mutations are associated with solid growth pattern and tumor-infiltrating leukocytes in lung adenocarcinoma. Mod Pathol 2013;26:1307-19. [Crossref] [PubMed]
- Yoshizawa A, Motoi N, Riely GJ, et al. Impact of proposed IASLC/ATS/ERS classification of lung adenocarcinoma: prognostic subgroups and implications for further revision of staging based on analysis of 514 stage I cases. Mod Pathol 2011;24:653-64. [Crossref] [PubMed]
- Ujiie H, Kadota K, Chaft JE, et al. Solid predominant histologic subtype in resected stage I lung adenocarcinoma is an independent predictor of early, extrathoracic, multisite recurrence and of poor postrecurrence survival. J Clin Oncol 2015;33:2877-84. [Crossref] [PubMed]
- Yanagawa N, Shiono S, Abiko M, et al. The clinical impact of solid and micropapillary patterns in resected lung adenocarcinoma. J Thorac Oncol 2016;11:1976-83. [Crossref] [PubMed]
- Wang Y, Zheng D, Zheng J, et al. Predictors of recurrence and survival of pathological T1N0M0 invasive adenocarcinoma following lobectomy. J Cancer Res Clin Oncol 2018;144:1015-23. [Crossref] [PubMed]
- Warth A, Penzel R, Lindenmaier H, et al. EGFR, KRAS, BRAF and ALK gene alterations in lung adenocarcinomas: Patient outcome, interplay with morphology and immunophenotype. Eur Respir J 2014;43:872-83. [Crossref] [PubMed]
- AMG 510 first to inhibit "Undruggable" KRAS. Cancer Discov 2019;9:988-9. Available online: https://doi.org/
10.1158/2159-8290.cd-nb2019-073 - Takenaka T, Takenoyama M, Yamaguchi M, et al. Impact of the epidermal growth factor receptor mutation status on the post-recurrence survival of patients with surgically resected non-small-cell lung cancer. Eur J Cardiothorac Surg 2015;47:550-5. [Crossref] [PubMed]
- Ito M, Miyata Y, Hirano S, et al. Synchronicity of genetic variants between primary sites and metastatic lymph nodes, and prognostic impact in nodal metastatic lung adenocarcinoma. J Cancer Res Clin Oncol 2019;145:2325-33. [Crossref] [PubMed]
Cite this article as: Ito M, Miyata Y, Okada M. Prognostic impact of targetable genetic variants in resected adenocarcinoma of the lung: a narrative review and model proposal for precise evaluation. Precis Cancer Med 2020;3:19.

