
NTRK and NRG1 gene fusions in advanced non-small cell lung cancer (NSCLC)
Introduction
Gene rearrangements are one of the most commonly observed genetic aberrations found in solid tumors and have growing diagnostic and therapeutic relevance. These genetic events were initially identified in hematological malignancies; breakpoint cluster region/Abelson murine leukemia viral oncogene homolog (BCR-ABL) rearrangement in chronic myeloid leukemia was the most prominent example. However, over the past several years, a multitude of gene rearrangements have been described in various solid tumors, in part due to the increased access to highly advanced sequencing technologies (1). Furthermore, implementing effective targeted drugs in rationally-designed, molecularly-selected clinical trials has prompted the rapid approval of some of these agents after only phase I studies either in specific histologies (for example crizotinib for anaplastic lymphoma kinase (ALK) and c-ros protooncogene 1 (ROS1) rearrangements in non-small cell lung cancer (NSCLC) (2,3) or in tumor agnostic patients [such as larotrectinib and entrectinib in neurotrophic receptor tyrosine kinase (NTRK) rearranged tumors] (4).
During the last two decades, breakthrough discoveries in lung cancer biology launched a new era in advanced NSCLC with the rise of personalized medicine. Since the discovery of epidermal growth factor receptor (EGFR) mutations in 2004, the list of molecularly defined subgroups of patients that can derive benefit from targeted therapies has grown considerably and current international guidelines recommend molecular testing for all patients with a newly diagnosed locally advanced or metastatic non-squamous NSCLC for at least 5–8 biomarkers for optimal patient selection (5-9). The incredible story of ALK rearrangements in NSCLC with the approval of the first-in-class inhibitor, crizotinib, only 4 years after the identification of ALK fusions in NSCLC (10) prompted the search for other oncogenic rearrangements potentially exploitable with targeted therapies. Neuregulin-1 (NRG1) and NTRK fusions are two of the most recently discovered rearrangements in NSCLC and represent two brilliant examples of tumor agnostic biomarkers. Although relatively rare, these two genetic aberrations represent two clinically relevant subgroups of NSCLC that can derive benefit from targeted therapies. Here we provide a comprehensive overview of the biological and clinicopathological characteristics of NRG1- and NTRK-rearranged NSCLC and the available data on the therapeutic exploitation of these targets.
NRG1 fusion genes
The epidermal growth factor (EGF) family plays an important role in both carcinogenesis and resistance to targeted therapy in NSCLC. NRGs are a group of growth factors for EGF receptor (11). The NRGs family of genes comprises four members (NRG1, NRG2, NRG3 and NRG4), and NRG1 is the most well studied. NRG1 has an essential role in normal physiology of the nervous system, heart and breast, in addition to a pathologic role in some diseases, including cancer (12). NRG1 presents three major isoforms, type I (heregulin), type II [glial growth factor-2 (GGF2)] and type III [sensory and motor neuron-derived factor (SMDF)], and six minor isoforms with specific function and expression (13).
All NRGs serve as ligands for the human epidermal growth factor receptor 4 (HER4), while NRG1/2 binds also for HER3 and are synthesized as transmembrane molecules. In addition, they can also act as soluble ligands after their release by membrane metalloproteases of the ADAM (a disintegrin and metalloproteinase) subfamily, such as tumor necrosis factor-alpha converting enzyme (TACE/ADAM17). NRGs binding, in an autocrine and juxtacrine manner, causes a conformational change of HER3, with exposition of the dimerization arm which can interact with other HER receptors, especially with HER2, leading to activation of the intracellular signaling cascade influencing critical cell processes such as growth and proliferation (14-16). Although HER3 lacks significant tyrosine kinase activity, its activation by NRGs is important in carcinogenesis by promoting heterodimerization with other HER receptors, leading to inappropriate activation; disrupting the interaction of NRGs with HER3 and HER4 receptors may be a potential therapeutic target (Figure 1).
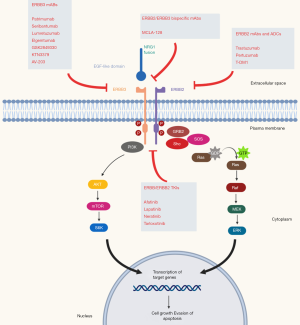
Beyond its role in carcinogenesis, NRGs are also involved in resistance to targeted therapy. Zhou and colleagues observed in NSCLC cell lines resistant to gefitinib that a selective ADAM 17 inhibitor (INCB3619) was able to reverse resistance to gefitinib, highlighting the contribution of NRG-dependent HER3 activation that contributes to gefitinib insensitivity in NSCLC (17). Upregulation of NRG1 has also been described as a mechanism of resistance to ALK inhibitors in NSCLC and systemic therapy in melanomas and HER2 inhibitor in breast cancer (18,19) and NRG1 mutations have been described in genetic disorders, such as the Hirschsprung disease.
To date, 18 different fusion partners for NRG1 have been reported in NSCLCs, although this list is destined to grow as many other variants have been described in other solid tumors (20-22). The CD74-NRG1 fusion variant is the most common in NSCLC and was first described in 2014 by Fernandez-Cuesta et al. (23). It consists of the first six exons of CD74 linked to the exons encoding the EGF-like domain β of the NRG1 isoform III (23). This gene fusion leads to extracellular expression of the EGF-like domain of NRG1 III-β3, which interacts with EGF receptors enabling hetero-dimerization of HER2-HER3 and subsequent activation. In lung cancer cell lines, ectopic expression of CD74-NRG1 showed a potential to promote cell tumor proliferation by activation of PI3K-AKT pathway through HER2 and HER3 receptors (23). This gene fusion seems particularly associated with a rare histological subtype that has been described in the 2015 WHO Classification of Lung Tumors (24), known as invasive mucinous adenocarcinoma (IMA) (23,25-27), which accounts for approximately 5% of all lung adenocarcinomas and harbors KRAS mutations in ~40–60% of the cases (25,28). IMA is associated with a poorer prognosis than other common subtypes of lung adenocarcinoma, including lepidic and acinar subtypes, and is rarely associated with EGFR mutations and ALK rearrangements (25,29,30).
In an analysis of 21,858 solid tumor specimens analyzed using RNA sequencing, 41 NRG1 fusions were identified. CD74-NRG1 was the most common variant (29%), followed by AT1P1-NRG1 (10%) and SDC4-NRG1 (7%) (20). Other fusion partners have been described at lower frequency and include TNC, MDK, DIP2B, KIF13B, RBPMS, MRPL13, ROCK1, DPYSL2, SLC3A2, VAMP2, WRN, ITGB1 and PARP8 in NSCLC (20-22,25,27,31,32). As commonly observed in other oncogene-addicted tumors, NRG1 fusions are usually mutually exclusive with other oncogenic drivers, such as EGFR, BRAF, and KRAS mutations or ALK, ROS1, and RET rearrangements (20). However, in selected cases, NRG1 fusions can coexist with other oncogenic drivers, such as ALK rearrangements (27,33) and KRAS amplification/mutations (27,32,34).
The reported incidence of NRG1 fusions is of ~0.2–0.5% in unselected NSCLCs (35,36) (Table 1).
Table 1
| Ref. | Ethnicity | Cohort | NRG1 fusions (%) | Concomitant oncogene drivers | Detection methods |
|---|---|---|---|---|---|
| (34) | Caucasian | 51 IMAs and 34 non-IMA cases | 31% IMAs and 3% non-IMAs | 11% (KRAS mutations) | FISH and RNA sequencing |
| (23) | Asian | 102 pan-negative LUAD NS | 3.9% (27% IMAs) | None | RT-PCR |
| (36) | Asian | 4,874 non-SqCC NSCLC | 0.3% | None | RT-PCR and NGS |
| (20) | NR | 9,592 NSCLC | 0.3% | None | NGS RNA-sequencing |
| (25) | Asian | 34 KRAS-negative IMAs | 17.6% | None | NGS RNA-sequencing |
| (26) | Asian | 13 IMAs | 8% | None | Direct RNA-sequencing |
| (27) | Asian | 59 IMAs | 27% | 62% (KRAS mutations) | NGS RNA-sequencing |
| (35) | Caucasian | 404 NSCLC | 0.5% | None | NGS RNA sequencing |
| (22) | Asian | 1681 LUAD | 0.36% | None | NGS RNA sequencing |
| (37) | Caucasian and Asian patients | 25 IMAs | 4% | None | FISH |
| (21) | Caucasian | 2,079 LUAD | 1.14% | None | NGS DNA-sequencing |
NRG1, neuregulin-1; LUAD, lung adenocarcinoma; IMA, invasive mucinous adenocarcinoma; NS, never smoker; non-SqCC, non-squamous; RT-PCR, reverse transcriptase polymerase chain reaction; NGS, next generation sequencing; NSCLC, non-small cell lung cancer; NR, not reported.
The gold standard for detection of NRG1 gene fusions is RNA sequencing in comparison with DNA sequencing, although fluorescence in situ hybridization (FISH) can be used as a pre-screening method for its detection, but only genetic sequencing will allow the identification of the gene fusion (13). In a recent multicenter registry of 117 NRG1-positive lung cancers, RNA-based assays (anchored multiplex PCR, nCounter, RT-PCR, and transcriptome) were the most common detection methods (79.5%), followed by FISH (12%) and DNA-based methods (hybrid capture-based NGS and amplicon-based NGS) (9.4%) (38). RNA-sequencing is associated with higher sensitivity for genetic rearrangements and can increase the detection of NRG1 gene fusions compared with DNA-based methods, which often do not cover the large introns in NRG1 (21).
The clinicopathological characteristics of NRG1 fusion-positive NSCLCs were recently analyzed in a large multicenter retrospective study evaluating 117 patients. NRG1 fusions were more frequently associated with female sex (54.7%) and never smoking history (43.6%), adenocarcinoma histology (94.9%), mostly of mucinous subtype (71%), and lung metastases (80% in stage IV patients). Treatment with platinum-based chemotherapy in 18 evaluable patients was associated with an 11% overall response rate (ORR) and a 61% disease control rate (DCR), whereas no responses were observed in patients treated with PD-1/PD-L1 inhibitors either as monotherapy (n=6) or in combination with chemotherapy (n=5) (38). This data, albeit limited by the small sample size, suggests that NRG1 fusion-positive NSCLCs, consistent with other oncogene-addicted subgroups, is associated with low response to immune checkpoint inhibitors and other treatment strategies should be pursued.
Preclinical studies demonstrated that NRG1 signals through induction of HER2–HER3 heterodimers, leading to subsequent PI3K-AKT pathway activation and stimulation of oncogenic growth, and that the downstream signaling is inhibited by HER2/HER3 blockage (23,39). Different HER2/HER3 inhibitors are approved in other clinical indications (such as afatinib, pertuzumab, and neratinib) or are under active clinical development. Several case reports and small retrospective studies provided clinical evidence of activity of these agents (Table 2).
Table 2
| Age (yrs) | Sex | Smoking habits | Histology | NRG1 Fusion | Treatment | Line(s) of therapy | Response | PFS (mos) | Treatment duration (mos) | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|
| Case reports with HER2/HER3 inhibitors | ||||||||||
| 43 | F | Never smoker | Adenocarcinoma | SDC4-NRG1 | Afatinib | 3rd | PR | 12 | 12 | (40) |
| 62 | F | Never smoker | IMA | CD74-NRG1 | Afatinib | 2nd | PR | 6 | 6 | (41) |
| 42 | M | Never smoker | Adenocarcinoma | SLC3A2-NRG1 | Afatinib | 2nd | PR | 12 | 18 | (35) |
| 62 | M | Never smoker | Mucinous adenocarcinoma | CD74-NRG1 | Afatinib | 1st | PR | 10 | >10 | (35) |
| 70 | F | Never smoker | Non-mucinous adenocarcinoma | NR | Afatinib | 15th | PR | 24 | 24 | (42) |
| 66 | F | Never smoker | Non-mucinous adenocarcinoma | CD74-NRG1 | Afatinib | 5th | PR | 19+ | 19+ | (42) |
| 68 | M | Former smoker | Non-mucinous adenocarcinoma | SDC4-NRG1 | Afatinib | 3rd | SD | 4 | 4 | (42) |
| 43 | F | Never Smoker | IMA | CD74-NRG1 | Afatinib | 4th | PR | 18+ | 18+ | (42) |
| 81 | M | Former cigar use | IMA | CD74-NRG1 | Afatinib | 1st | SD | 3 | 3 | (21) |
| 52 | F | Current smoker | IMA | SDC4-NRG1 | Afatinib | 2nd | PD | 1 | 1 | (21) |
| 51 | M | Former smoker | IMA | CD74-NRG1 | Afatinib | 1st | PD | 2 | 2 | (21) |
| 86 | M | N.R. | IMA | CD74-NRG1 | GSK2849330 | 5th | PR | 19 | N.R. | (21) |
| 55 | F | Never Smoker | IMA | SLC3A2-NRG1 | Lumretuzumab + erlotinib | 6th | SD | 3.8 | 3.8 | (43) |
| 42 | F | Never Smoker | IMA | SLC3A2-NRG1 | Lumretuzumab + erlotinib | 6th | SD | 3.8 | 3.8 | (43) |
| Retrospective studies with HER2/HER3 inhibitors in NRG1 gene fusions positive NSCLC | ||||||||||
| 12 (number of pts) | Afatinib* | 1–15 | ORR 33%, DCR 50% | PFS 2.0 months, OS not reached | (38) | |||||
*, one patient was treated with afatinib in combination with docetaxel-ramucirumab. NRG1, neuregulin-1; NR, not reported; mos, months; yrs, years; ORR, overall response rate; DCR, disease control rate; PFS, progression free survival; OS, overall survival; pts, patients; IMA, invasive mucinous adenocarcinoma; NSCLC, non-small cell lung cancer; PR, partial response; SD, stable disease; PD, progressive disease.
Afatinib is a potent and selective pan-inhibitor of HER family blocker, that covalently binds to and irreversibly blocks signaling from all homo- and heterodimers formed by the HER family members, including EGFR (HER1), HER2, HER3 and HER4. It is currently Food and Drug Administration (FDA) approved for the treatment of EGFR mutated NSCLCs based on the results of the phase III trials LUX-Lung-3 and -6 (44,45) and for the treatment of squamous cell lung cancer after prior platinum-based chemotherapy based on the results of the LUX-Lung-8 study (46). A growing body of evidence suggest that afatinib is a potential treatment option for patients with NRG1 fusion-positive tumors across multiple cancer types, including NSCLC, as reported in multiple case reports (Table 2). Recently a multicenter global registry of 117 NRG1-positive cases described afatinib activity in 12 patients with stage IV NSCLC harboring an NRG1 gene fusion. In these heavily pretreated patients (line of treatment ranging from 1 to 15), afatinib was associated with a 33% ORR, a 50% DCR and a median PFS of 2.0 months. In a few patients, long term responses to afatinib were observed. However, the presence of NRG1 gene fusion was associated with a favorable OS (4.83 months in stage IV patients) and was not significantly influenced by treatment with afatinib (38). Prospective studies are ongoing in the Drug Rediscovery Protocol trial (DRUP) (NCT02925234) and the Targeted Agent and Profiling Utilization Registry study (TAPUR) (NCT02693535) (42).
Early clinical proof-of-principle data demonstrated activity with HER3-directed targeted therapy in patients with advanced NRG1-rearranged cancers. GSK2849330 is an HER3 monoclonal antibody (mAb) that blocks the binding of NRG1 to HER3 and inhibits receptor heterodimerization. Drilon et al. reported a dramatic (32% tumor reduction) and durable response (19 months) to this HER3 mAb in an 86-year-old male with IMA harboring a CD74-NRG1 gene fusion enrolled in an NSCLC expansion cohort of a phase I trial (NCT01966445). This trial included more 28 patients with similar and higher HER3 expression, but without NRG1 gene fusions and none of these patients responded to therapy. This clinical data is supported by preclinical evidence of antiproliferative activity in NRG1 fusion-positive cell line MDA-MB-175-VII and durable tumor regression in a patient-derived xenograft (PDX) mouse model (21). In contrast, afatinib was associated with a significant reduction in tumor growth compared with vehicle, but no tumor regression was observed in the PDX model and no responses were observed in three patients harboring NRG1 rearrangements (21). Several other HER3 inhibitors have been evaluated in clinical trials, such as patritumab (U3-1287, AMG-888), seribantumab (MM-121/SAR256212), lumretuzumab (RG7116), AV-203 and elgemtumab (LJM 716), these studies did not focus on NRG1 gene fusions. The HER2/HER3 bispecific antibody MCLA-128, which blocks both NRG1 binding and HER2/HER3 heterodimerization, showed potent in vitro and in vivo activity in NRG1 fusion-positive models (47). A global phase II basket trial (NCT02912949) is ongoing and will evaluate the safety and activity of this compound in three NRG1 fusion-positive cohorts: pancreatic cancer (n=25), NSCLC (n=25), and other solid tumors (n=40). NRG1 gene fusion assessment can be done with different molecular assay such as PCR, NGS (RNA orDNA) or FISH (48).
NTRK fusion genes
NTRK genes encode tropomyosin receptor kinases (Trk) family proteins, which includes three members (TRKA, TRKB and TRKB, encoded by NTRK1, NTRK2, and NTRK3, respectively). These receptor tyrosine kinases play a physiologic role in central and peripheral nervous system development and are activated by different ligands, including nerve growth factor (NGF), brain-derived neurotrophic factor (BDNF), neurotrophin 4 (NT-4), and neurotrophin 3 (NT-3) (49-51). Once activated, TRKs signal through three main downstream pathways (MAPK, PI3K and PLC-γ), resulting in neuronal development and differentiation (52) (Figure 2).
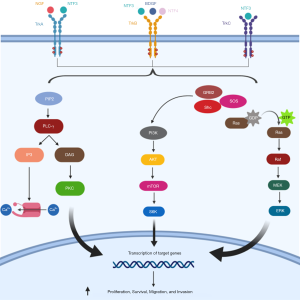
Different mechanisms can be responsible of TRK oncogenic activation, although gene fusions involving NTRK1, NTRK2 or NTRK3 are the most commonly observed in solid tumors, including NSCLC. Several gene partners have been described to date and the majority harbor oligomerization domains that can constitutively activate the kinase domain of TRK (51).
NTRK gene fusions were first identified in NSCLC in 2013 by Vaishnavi et al. through a targeted DNA NGS assay on tumor samples from 36 lung cancer patients without known oncogenic alterations, identifying two NTRK1 gene fusions with MPRIP and CD74, respectively. They also developed a FISH assay to detect NTRK1 rearrangements, reporting another additional case in a cohort of 56 lung adenocarcinoma samples without detectable oncogenic alterations (overall frequency 3.3%) (50). Since this initial report, subsequent studies have identified NTRK gene fusions in NSCLC with a frequency <1% in unselected patients (Table 3).
Table 3
| Study | Population [n] | Frequency (%) | NTRK | Fusion partner(s) | Detection methods |
|---|---|---|---|---|---|
| Farago, 2018 (53) | NSCLC [4,872] | 0.23% | NTRK1, NTRK3 | SQSTM1, TPR, IRF2BP2, TM3, MPRIP, ETV6 | DNA NGS, RNA NGS or FISH |
| Vaishnavi, 2013 (50) | LUAD without oncogenic drivers [91] | 3.3% | NTRK1 | MPRIP, CD74 | DNA NGS or FISH |
| Stransky, 2014 (54) | LUAD [513] | 0.19% | NTRK2 | TRIM24 | RNA sequencing |
| Miyamoto, 2019 (36) | Non-SqCC NSCLC [4,874] | 0.05% | NTRK3 | NR | RT-PCR and NGS |
| Gatalica, 2018 (55) | LUAD [4,073] | 0.1% | NTRK1-3 | TPM3, SQSTM1, ETV6 | DNA and RNA NGS & IHC |
| Ou, 2019 (56) | NSCLC [42,791] | 0.1% | NTRK1-3 | IRF2BP2, TPM3, and others | DNA NGS |
| Xia, 2019 (57) | NSCLC [21,155] | 0.056% | NTRK1 | CD74, IRF2BP2, LMNA, PHF20, SQSTM1, TPM3, TRP | DNA NGS |
NTRK, neurotrophic receptor tyrosine kinase; NSCLC, non-small cell lung cancer; LUAD, lung adenocarcinoma; non-SqCC, non-squamous; RT-PCR, reverse transcriptase polymerase chain reaction; NGS, next generation sequencing; NR, not reported.
In contrast to other oncogenic rearrangements, such as ALK and ROS1 translocations, that are associated with peculiar clinic-pathological characteristics (58,59), NTRK gene fusions seem not limited to specific subgroups of NSCLC patients and can occur in both squamous and non-squamous histology, including neuroendocrine carcinomas, independently of sex and smoking status (53). A large retrospective study analyzed data from 166,067 real world solid tumor samples sequenced by Foundation Medicine (FMI), showing that NTRK gene fusions do not co-occur with clinically actionable drivers in solid tumors, present a tumor mutational burden (TMB) generally similar to NTRK fusion-negative solid tumors, and occur at a slightly higher frequency in patients with Asian ancestry (0.46% in East Asian, 0.37% in South Asian, 0.34% in American, 0.29% in European and 0.32% in African) (60). Furthermore, NTRK gene fusions in NSCLC seemed associated with levels of TMB and frequencies of PD-L1 expression higher than other molecularly defined subgroups (EGFR, ALK and ROS1 altered cases) and co-exist with STK11 mutations, which have been associated with decreased efficacy to immune checkpoint inhibitors (61), at frequencies similar to NSCLC in general but lower than the frequency in lung adenocarcinoma only (56). This data suggests that NTRK fusion positive NSCLCs might benefit from immunotherapy than is usually associated with lower efficacy in other oncogene addicted NSCLC subgroups (62). A recent retrospective study in a Chinese population showed that NTRK1 fusions may coexist with EGFR mutations in EGFR tyrosine kinase inhibitor (TKI)-pretreated patients and might represent a potential mechanism of acquired resistance to these agents (57).
Besides NSCLC, NTRK fusions have been found in multiple tumors types that can be grouped according to the frequency at which these fusions are detected in:
- Rare cancer types that present NTRK fusions with a prevalence >90%, including secretory breast carcinoma, mammary analogue secretory carcinoma (MASC), congenital mesoblastic nephroma (cellular or mixed subtypes) and infantile fibrosarcomas;
- Common solid tumors with a prevalence of NRTK fusions of 5–25%, such as papillary thyroid cancers, spitzoid neoplasms, gastrointestinal stromal tumors (GIST) lacking canonical KIT, PDGFRA or RAS alterations, and certain pediatric gliomas;
- Common solid tumors with low NTRK gene fusions prevalence (<5%, but predominantly <1%), such as lung or pancreatic adenocarcinomas, head and neck squamous cell, biliary duct, breast, colorectal and renal cell carcinomas, melanomas, primary brain tumors of adulthood (such as astrocytomas or glioblastomas) and non-GIST soft-tissue sarcomas (51,52).
Different detection methods have been reported to date for NTRK fusions, including immunohistochemistry (IHC), FISH, reverse transcription PCR (RT-PCR), DNA-based NGS, RNA-based NGS, and DNA/RNA hybrid sequencing assays. Each of these methodologies is associated with different sensitivity and specificity for NTRK fusions as well as different turnaround time and cost (63). Selection of the appropriate assay for NTRK fusion detection seems to be influenced by tumor type and genes involved, as well as other factors such as available material, accessibility of various clinical assays, and whether comprehensive genomic testing is needed concurrently. Indeed, a recent retrospective analysis of 87 patients with oncogenic NTRK1-3 fusions with various solid tumors identified by a targeted DNA-based NGS (MSK-IMPACT) or an RNA-based sequencing assay (MSK-Fusion) were tested with pan-Trk IHC. DNA-based sequencing showed an overall sensitivity and specificity of 81.1% and 99.9%, respectively, for the detection of NTRK fusions compared to RNA-based sequencing, where false negatives occurred when fusions involved breakpoints not covered by the assay. IHC showed overall sensitivity of 87.9% and specificity of 81.1%. Sensitivity was different according to the fusion type (96% for NTRK1, 100% for NTRK2 fusions, and 79% for NTRK3 fusions) and specificity differed by tumor histology (100% for carcinomas of the colon, lung, thyroid, pancreas, and biliary tract, but decreased to 82% and 52% for breast and salivary gland carcinomas, respectively) (64). A reasonable approach is to consider FISH or, if not available, pan-Trk IHC as the diagnostic test for rare tumors with high NTRK fusion prevalence (>90%), such as mammary analogue secretory carcinoma, congenital mesoblastic nephroma, infantile fibrosarcoma or secretory breast carcinoma. NGS confirmation of pan-Trk IHC positive cases can be conducted concurrently with treatment decision and should be considered in FISH/IHC negative cases. For tumors with lower frequency of NTRK fusions (5–25%) or rarely associated with these oncogenic drivers (<5%), such as lung cancer, the diagnostic algorithm depends on the use of NGS as diagnostic tool in routine clinical practice. If NGS is routinely performed for molecular testing, NTRK fusions should be incorporated in NGS analysis. Alternatively, if NGS is not routinely performed for that specific tumor histology type or institutional unavailability, pan-Trk IHC can be used as screening test, followed by confirmatory NGS in positive cases (65). Similarly, the European Society for Medical Oncology (ESMO) recommendations for NTRK testing incorporated the use of FISH, RT-PCR or targeted RNA NGS assays for solid tumors known to harbor highly recurrent NTRK fusions, while upfront use of NGS (preferably RNA-based) followed by IHC to confirm positive cases or alternatively IHC as screening tool followed by NGS for tumors harboring NTRK fusions with lower frequency, as NSCLC (66).
Several TKIs with various degrees of activity against TRKA, TRKB and/or TRKC have been developed and two (larotrectinib and entrectinib) have been recently approved by the US FDA.
Larotrectinib (also known as LOXO-101 and ARRY-470) is a potent and highly selective pan-TRK (TRKA, TRKB, and TRKC) ATP-competitive inhibitor with a >100-fold selectivity for inhibition of TRK versus other kinases and >1,000-fold selectivity for tested non-kinase targets. In addition, larotrectinib was designed to have limited central nervous system penetration to reduce the potential risk of neurological toxicity due to the inhibition of TRK receptors normally expressed in the brain (67). The development program of larotrectinib in NTRK fusion-positive tumors included three studies: a phase I study involving adults, a phase I/II study involving children, and a phase II basket trial involving adolescents and adults (NAVIGATE). The preliminary analysis of the first 55 patients with 17 unique TRK fusion-positive tumor types (including 7% lung cancer patients) enrolled in the phase I studies reported a 75% ORR by independent central review (80% per investigators) and an 80% DCR. The recommended phase II dose (RP2D) was 100 mg twice daily for adults and children with a body surface area (BSA) ≥1 m2 of and 100 mg/m2 for children with a BSA <1 m2 (4). Larotrectinib was well tolerated with adverse events (AE) mainly of grade 1–2, with 13% of patients developing a grade 3-4 event and only one patient discontinued due to an AE related to larotrectinib (68). Based on this preliminary data, larotrectinib was the first in class highly selective pan-TRK inhibitor to gain FDA approval and European Medicine Agency (EMA) conditional approval, independently of tumor histology.
Updated data of this primary cohort with additional 98 patients (153 in total) enrolled in an expanded patient cohort were recently presented at the 2019 ESMO meeting (68), confirming the impressive activity of larotrectinib [79% ORR, 95% confidence interval (CI): 72–85%] in NTRK fusion-positive patients. Responses were durable with a median duration of response of 35.2 months (95% CI: 22.8–NE) and a reported median PFS of 28.3 months (95% CI, 22.1–NE) and a median OS of 44.4 months (95% CI, 36.5–NE) in the integrated dataset of the expanded cohort (n=159, including 12 lung cancer patients) (68). Intracranial activity of larotrectinib has been reported recently in two NTRK fusion-positive patients with lung adenocarcinoma and triple negative breast cancer enrolled in the NAVIGATE study (69).
Preliminary data of the mechanisms of resistance to larotrectinib have also been reported in the primary cohort. Interestingly, of six patients with primary resistance, one had been pretreated with entrectinib and harbored a NTRK3 G623R mutation, which is associated with steric interference of drug binding, and three of five had an unconfirmed TRK IHC expression. Furthermore, preliminary data of the mechanisms of acquired resistance to larotrectinib were reported, including substitution in the solvent front position (NTRK1 G595R and NTRK3 G623R mutations) or in the gatekeeper position (NTRK1 F589L mutations) or the xDFG (a portion of the kinase-activation loop) position (NTRK1 G667S and NTRK3 G696A mutations) (4). In order to overcome acquired resistance mediated by recurrent kinase domain (solvent front and xDFG) mutations, a next generation TRK inhibitor, known as LOXO-195 (BAY 2731954), was designed. This compound demonstrated potent and selective activity against all three TRK kinases, their fusions, and acquired resistance mutations both preclinically and in patients (70). Preliminary safety and efficacy data of the phase I study (NCT03215511, n=20) and the FDA expanded access single patient protocol (n=11) were recently presented. LOXO-195 reported an ORR of 34% with a promising 45% ORR in 20 patients with TRK kinase mutations (50% in both solvent front and xDFG mutations and 25% in gatekeeper mutations) and lower ORR among those with unknown mechanisms of resistance (17%) and with by-pass track mechanism (0%) (71).
Entrectinib (RXDX-101, NMS-E628) is a pan-TRK, ROS1 and ALK ATP-competitive inhibitor with ability to cross the blood-brain-barrier (BBB) (72) that was recently FDA approved the treatment of patients with metastatic or unresectable solid tumors harboring a NTRK gene fusion without a known acquired resistance mutation and also for the treatment of metastatic ROS1 positive NSCLC. The combined analysis of two phase I trials of entrectinib (ALKA-372-001 and STARTRK-1) in 119 patients with various solid tumors, including 71 NSCLC (60%), reported a relatively safe toxic profile with the majority of AEs grade 1–2 and dose reduction required in only 15% of the patients. The RP2D was 600 mg daily. Preliminary data of efficacy were reported. No responses were observed in patients without genetic rearrangements of NTRK1-3, ROS1 or ALK, with the exception of one patient with an ALK F124SV mutant neuroblastoma, and in ROS1/ALK fusion-positive patients who had been pretreated with one or more previous TKIs. The analysis of patients harboring NTRK1-3, ROS1 or ALK rearrangements and no previous TKIs (“phase II eligible population”, n=25) showed a 100% ORR in three NTRK fusion positive patients with measurable disease, including one NSCLC, and a 60% disease reduction in an additional patient with a glioneuronal tumor. Promising activity was also seen in TKI-naïve ROS1 (ORR 86%) and ALK (ORR 57%) rearranged tumors (73). An updated integrated analysis of phase I/II studies with entrectinib (ALKA-372-001, STARTRK-1, and STARTRK-2) was recently presented at the 2019 ESMO annual meeting (74,75). Among 54 NTRK fusion positive solid tumors, entrectinib reported a 59.3% ORR by blinded independent central review with a median duration of response of 12.9 months and a median OS of 23.9 months (95% CI, 16.8–NE). As expected, entrectinib was highly active even in patients with baseline brain metastases with an intracranial ORR of 54.5% and median intracranial duration of response not reached (75). The results in the NSCLC cohort (10 NTRK fusion positive patients) were consistent with the overall population with a 70% ORR and 10% complete response (74). Despite deep and clinically meaningful responses in many patients, resistance to entrectinib eventually occurs. The mechanisms of resistance were recently investigated in plasma samples from NTRK and ROS1 fusion positive patients enrolled in the phase II basket trial STARTRK-2 using a plasma NGS platform (Foundation Medicine). Acquired resistance mutations were detected in 34% and 28% of NTRK and ROS1 fusion positive patients, respectively, and off-target mechanisms of acquired resistance within the MAPK pathway were also reported in both groups (76). These data are in line with a recent report evaluating the mechanisms of resistance to various TRK inhibitors using tumor biopsies and cell free DNA and showing that MAPK signaling activation is a recurrent and convergent by-pass mediated resistance mechanism to various TRK inhibitors, including both first generation (larotrectinib and entrectinib) and next generation (LOXO195). The combination of TRK and MEK inhibitors has been shown to overcome these resistance mechanisms and, given the non-overlapping toxicities, might represent a promising therapeutic strategy in NTRK fusion positive patients (77).
Repotrectinib (TPX-0005) is next generation TRK, ALK and ROS1 TKI rationally designed to inhibit solvent-front substitutions (such as ALK G1202R, ROS1 G2032R or ROS1 D2033N, and TRKA G595R).
Repotrectinib exhibits activity against a variety of solvent front substitutions in vitro and in vivo and showed preliminary activity in patients with advanced ALK, ROS1, or NTRK1–3-rearranged cancers in the first-in-human dose-escalation phase I/II clinical trial (TRIDENT-1, NCT03093116), including a patient with a mammary analogue secretory carcinoma harboring an ETV6-NTRK3 rearrangement and a NTRK3 G623E mutation acquired after progressing to entrectinib (78). Preliminary safety data on first 83 patients treated with various repotrectinib doses (from 40 mg daily to 200 mg twice a day under fasted/feed conditions) showed a relatively safe toxicity profile. Most AEs were manageable and grade 1–2. The most common treatment-emergent AEs were dizziness (57%), dysgeusia (51%), dyspnea (30%), and fatigue (30%). Four dose-limiting toxicities occurred and were manageable with dose modifications: dyspnea/hypoxia G3 (n=1); G2 (n=1) and G3 (n=1) dizziness at 160 mg BID, and G3 dizziness (n=1) at 240 mg QD (79). The study is ongoing and efficacy data on NTRK fusion-positive patients are eagerly awaited.
Other TRK inhibitors that showed preclinical activity in NTRK fusion-positive models include the new selective ROS1/NTRK inhibitor DS-6051b (80), the IGF-1R/NTRK inhibitor BMS-536924 (81), the dual ALK/NTRK inhibitor TSR-011 (82), the multikinase inhibitor merestinib (LY2801653) (83), and the MET/TRK inhibitor altiratinib (DCC-2701) (84). Ongoing clinical trials with TRK inhibitors in NTRK fusion-positive solid tumor are summarized in Table 4.
Table 4
| Study name | Phase | Drug | Population | Estimated enrollment (n) |
|---|---|---|---|---|
| NAVIGATE (NCT02576431) | II | Larotrectinib | Adults and children with NTRK-fusion positive solid tumors | 320 patients |
| STARTRK-2 (NCT02568267) | II | Entrectinib | NTRK-, ROS1- and ALK-fusion positive solid tumors | 300 patients |
| NCT01639508 | I | Cabozantinib | NTRK fusion, or MET or AXL overexpression, amplification, or mutation (group B) | 68 patients (groups A-C) |
| NCT03215511 | I/II | LOXO-195 | Adult and pediatric subjects with previously treated NTRK fusion cancers | 93 patients |
| TRIDENT-1 (NCT03093116) | I/II | Repotrectinib | NTRK-, ROS1- and ALK-fusion positive solid tumors | 450 patients |
| NCT02675491 | I | DS-6051b | NTRK- or ROS1-fusion positive solid tumors | 15 patients |
| NCT01804530 | I | PLX7486 | Solid tumors, including NTRK-fusion positive | 59 patients-discontinued |
| NCT02920996 | II | Merestinib | NTRK-fusion positive solid tumors or MET-mutation NSCLC | 25 patients |
| NCT03556228 | I | VMD-928 | NTRK1 alterations, including fusions, positive solid tumors | 54 patients |
| NCT02219711 | I | MGCD516 | NTRK-fusion positive NSCLC | 260 patients |
| ONTRK (NCT03182257) | I | ONO-7579 | NTRK-fusion positive solid tumors | 1 patient enrolled-discontinued due to commercial reasons |
NTRK, neurotrophic receptor tyrosine kinase; NSCLC, non-small cell lung cancer.
Conclusions and future perspectives
Personalized medicine has revolutionized the therapeutic approach to most of the solid tumors, including NSCLC, with unprecedented results in molecularly defined patient subgroups. This led to a dramatic shift in our vision of this disease moving from the old concept of a unique, highly frequent, indistinct entity to a multitude of several different molecular entities with peculiar clinico-pathological and therapeutic characteristics. Recently, the identification of rare genetic rearrangements at low frequency in different solid tumors has changed the old vision of drug development leading to the approval of targeted therapies after only phase I studies and independently of tumor histology. NTRK and NRG1 gene fusions represent two of the most compelling examples of tumor agnostic biomarkers and, although present at very low frequency in NSCLC, constitute two clinically relevant subgroups of patients that can derive benefit from matched targeted drugs. The approval of the TRK inhibitors larotrectinib and entrectinib and the rapid clinical development of next generation TRK TKIs is reshaping the therapeutic algorithm of this small subgroups of patients, adding NTRK gene fusions to the list of genes that should be tested for the optimal treatment selection of NSCLC. The availability of highly potent and selective drugs directed against NTRK rearrangements further reinforce the utility of multiplex molecular testing in NSCLC overcoming the limits of single gene tests. NRG1 fusion is the latest oncogene driver that has shown promising pharmacological exploitation, although the optimal therapeutic sequence should be still defined. Ongoing clinical trials with pan-HER TKIs (afatinib) or bispecific monoclonal antibodies targeting HER2/HER3 agents, such as MCLA-128, will provide definitive conclusions on the activity of targeted therapies in this rare subgroup of patients.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Alfredo Addeo and Giuseppe Banna) for the Series “Non-Small Cell Lung Cancer” published in Precision Cancer Medicine. The article was sent for external peer review organized by the Guest Editors and the editorial office.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/pcm.2020.03.02). The Series “Non-Small Cell Lung Cancer” was commissioned by the editorial office without any funding or sponsorship. RM reports grants from Astra Zeneca, other from Genentech, outside the submitted work. SVL reports grants from Alkermes, grants from AstraZeneca, grants from Bayer, grants from Blueprint Medicines, grants from Bristol-Myers Squibb, grants from Merck/MSD, grants from Merus, grants from Molecular Partners, grants from Pfizer, grants from Rain Therapeutics, grants from RAPT Therapeutics, grants from Spectrum, grants from Takeda/ARIAD, grants from Turning Point Therapeutics, grants from Ignyta, grants from Clovis Oncology, grants from Debiopharm, grants from Esanex, grants from Genentech/Roche, grants from Eli Lilly, grants from Lycera, grants from Corvus Pharmaceuticals, personal fees from Apollomics, personal fees from AstraZeneca, personal fees from Bristol-Myers Squibb, personal fees from Celgene, personal fees from G1 Therapeutics, personal fees from Genentech/Roche, personal fees from Guardant Health, personal fees from Janssen, personal fees from Eli Lilly, personal fees from Loxo Oncology, personal fees from Merck/MSD, personal fees from Pfizer, personal fees from PharmaMar, personal fees from S.A., personal fees from Regeneron Pharmaceuticals, personal fees from Taiho Oncology, personal fees from Takeda/ARIAD, personal fees from Ignyta, personal fees from Tempus, non-financial support from AstraZeneca, non-financial support from Genentech/Roche, non-financial support from Merck/MSD, outside the submitted work. CR reports grants from MSD, grants from Astra Zeneca, grants from ARCHER, grants from Inivata, grants from Merck Serono, grants from Mylan, non-financial support from Oncopass, grants from Lung Cancer Research Foundation-Pfizer, non-financial support from Guardant Health, non-financial support from Biomark inc., outside the submitted work. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Disclosure: Dr. Rolfo reports advisor board role for Inivata, ARCHER, MD Serono, Oncompass (non-financial), and Mylan; speakers’ bureau for Astra Zeneca and MSD; honoraria from Elsevier; grants/research support from Lung Cancer Research Foundation (LCRF), American Cancer Society (ACS), GuardantHealth (non-financial), and Biomarkers (non-financial).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Schram AM, Chang MT, Jonsson P, et al. Fusions in solid tumours: diagnostic strategies, targeted therapy, and acquired resistance. Nat Rev Clin Oncol 2017;14:735-48. [Crossref] [PubMed]
- Kwak EL, Bang YJ, Camidge DR, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med 2010;363:1693-703. [Crossref] [PubMed]
- Shaw AT, Ou SHI, Bang YJ, et al. Crizotinib in ROS1-rearranged non-small-cell lung cancer. N Engl J Med 2014;371:1963-71. [Crossref] [PubMed]
- Drilon A, Laetsch TW, Kummar S, et al. Efficacy of Larotrectinib in TRK Fusion-Positive Cancers in Adults and Children. N Engl J Med 2018;378:731-9. [Crossref] [PubMed]
- Ettinger DS, Wood DE, Aggarwal C, et al. NCCN Clinical Practice Guidelines in Oncology - Non-Small Cell Lung Cancer, version 7.2019. Available online: https://www.nccn.org/professionals/physician_gls/pdf/nscl_blocks.pdf
- Planchard D, Popat S, Kerr K, et al. Metastatic non-small cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2019;30:863-70. [Crossref] [PubMed]
- Wu YL, Planchard D, Lu S, et al. Pan-Asian adapted Clinical Practice Guidelines for the management of patients with metastatic non-small-cell lung cancer: a CSCO-ESMO initiative endorsed by JSMO, KSMO, MOS, SSO and TOS. Ann Oncol 2019;30:171-210. [Crossref] [PubMed]
- Kalemkerian GP, Narula N, Kennedy EB, et al. Molecular Testing Guideline for the Selection of Patients With Lung Cancer for Treatment With Targeted Tyrosine Kinase Inhibitors: American Society of Clinical Oncology Endorsement of the College of American Pathologists/International Association for the Study of Lung Cancer/Association for Molecular Pathology Clinical Practice Guideline Update. J Clin Oncol 2018;36:911-9. [Crossref] [PubMed]
- Lindeman NI, Cagle PT, Aisner DL, et al. Updated Molecular Testing Guideline for the Selection of Lung Cancer Patients for Treatment With Targeted Tyrosine Kinase Inhibitors: Guideline From the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. J Thorac Oncol 2018;13:323-58. [Crossref] [PubMed]
- Blackhall F, Cappuzzo F. Crizotinib: from discovery to accelerated development to front-line treatment. Ann Oncol 2016;27:iii35-41. [Crossref] [PubMed]
- Gullick WJ. The epidermal growth factor system of ligands and receptors in cancer. Eur J Cancer 2009;45:205-10. [Crossref] [PubMed]
- Falls DL. Neuregulins: functions, forms, and signaling strategies. Exp Cell Res 2003;284:14-30. [Crossref] [PubMed]
- Nagasaka M, Ou SHI. Neuregulin 1 Fusion-Positive NSCLC. J Thorac Oncol 2019;14:1354-9. [Crossref] [PubMed]
- Montero JC, Rodriguez-Barrueco R, Ocana A, et al. Neuregulins and cancer. Clin Cancer Res 2008;14:3237-41. [Crossref] [PubMed]
- Jeong H, Kim J, Lee Y, et al. Neuregulin-1 induces cancer stem cell characteristics in breast cancer cell lines. Oncol Rep 2014;32:1218-24. [Crossref] [PubMed]
- Fernandez-Cuesta L, Thomas RK. Molecular Pathways: Targeting NRG1 Fusions in Lung Cancer. Clin Cancer Res 2015;21:1989-94. [Crossref] [PubMed]
- Zhou B-BS, Peyton M, He B, et al. Targeting ADAM-mediated ligand cleavage to inhibit HER3 and EGFR pathways in non-small cell lung cancer. Cancer Cell 2006;10:39-50. [Crossref] [PubMed]
- Cheng H, Terai M, Kageyama K, et al. Paracrine Effect of NRG1 and HGF Drives Resistance to MEK Inhibitors in Metastatic Uveal Melanoma. Cancer Res 2015;75:2737-48. [Crossref] [PubMed]
- Kimura M, Endo H, Inoue T, et al. Analysis of ERBB ligand-induced resistance mechanism to crizotinib by primary culture of lung adenocarcinoma with EML4-ALK fusion gene. J Thorac Oncol 2015;10:527-30. [Crossref] [PubMed]
- Jonna S, Feldman RA, Swensen J, et al. Detection of NRG1 Gene Fusions in Solid Tumors. Clin Cancer Res 2019;25:4966-72. [Crossref] [PubMed]
- Drilon A, Somwar R, Mangatt BP, et al. Response to ERBB3-Directed Targeted Therapy in NRG1-Rearranged Cancers. Cancer Discov 2018;8:686-95. [Crossref] [PubMed]
- Pan Y, Zhang Y, Ye T, et al. Detection of Novel NRG1, EGFR, and MET Fusions in Lung Adenocarcinomas in the Chinese Population. J Thorac Oncol 2019;14:2003-8. [Crossref] [PubMed]
- Fernandez-Cuesta L, Plenker D, Osada H, et al. CD74-NRG1 fusions in lung adenocarcinoma. Cancer Discov 2014;4:415-22. [Crossref] [PubMed]
- Travis WD, Brambilla E, Nicholson AG, et al. The 2015 World Health Organization Classification of Lung Tumors: Impact of Genetic, Clinical and Radiologic Advances Since the 2004 Classification. J Thorac Oncol 2015;10:1243-60. [Crossref] [PubMed]
- Nakaoku T, Tsuta K, Ichikawa H, et al. Druggable oncogene fusions in invasive mucinous lung adenocarcinoma. Clin Cancer Res 2014;20:3087-93. [Crossref] [PubMed]
- Gow CH, Wu SG, Chang YL, et al. Multidriver mutation analysis in pulmonary mucinous adenocarcinoma in Taiwan: identification of a rare CD74-NRG1 translocation case. Med Oncol 2014;31:34. [Crossref] [PubMed]
- Shin DH, Lee D, Hong DW, et al. Oncogenic function and clinical implications of SLC3A2-NRG1 fusion in invasive mucinous adenocarcinoma of the lung. Oncotarget 2016;7:69450-65. [Crossref] [PubMed]
- Maeda Y, Tsuchiya T, Hao H, et al. Kras(G12D) and Nkx2-1 haploinsufficiency induce mucinous adenocarcinoma of the lung. J Clin Invest 2012;122:4388-400. [Crossref] [PubMed]
- Sun F, Wang P, Zheng Y, et al. Diagnosis, clinicopathological characteristics and prognosis of pulmonary mucinous adenocarcinoma. Oncol Lett 2018;15:489-94. [PubMed]
- Tsuta K, Kawago M, Inoue E, et al. The utility of the proposed IASLC/ATS/ERS lung adenocarcinoma subtypes for disease prognosis and correlation of driver gene alterations. Lung Cancer 2013;81:371-6. [Crossref] [PubMed]
- Dhanasekaran SM, Balbin OA, Chen G, et al. Transcriptome meta-analysis of lung cancer reveals recurrent aberrations in NRG1 and Hippo pathway genes. Nat Commun 2014;5:5893. [Crossref] [PubMed]
- Xia D, Le LP, Iafrate AJ, et al. KIF13B-NRG1 Gene Fusion and KRAS Amplification in a Case of Natural Progression of Lung Cancer. Int J Surg Pathol 2017;25:238-40. [Crossref] [PubMed]
- Muscarella LA, Trombetta D, Fabrizio FP, et al. ALK and NRG1 Fusions Coexist in a Patient with Signet Ring Cell Lung Adenocarcinoma. J Thorac Oncol 2017;12:e161-3. [Crossref] [PubMed]
- Trombetta D, Graziano P, Scarpa A, et al. Frequent NRG1 fusions in Caucasian pulmonary mucinous adenocarcinoma predicted by Phospho-ErbB3 expression. Oncotarget 2018;9:9661-71. [Crossref] [PubMed]
- Gay ND, Wang Y, Beadling C, et al. Durable Response to Afatinib in Lung Adenocarcinoma Harboring NRG1 Gene Fusions. J Thorac Oncol 2017;12:e107-10. [Crossref] [PubMed]
- Miyamoto S, Matsumoto S, Yoh K, et al. 1481OClinical development of molecular-targeted therapies for non-small cell lung cancer through nationwide genome screening in Japan (LC-SCRUM-Japan). Ann Oncol 2019;30:mdz260.003.
- Duruisseaux M, McLeer-Florin A, Antoine M, et al. NRG1 fusion in a French cohort of invasive mucinous lung adenocarcinoma. Cancer Med 2016;5:3579-85. [Crossref] [PubMed]
- Duruisseaux M, Liu SV, Han JY, et al. NRG1 fusion-positive lung cancers: Clinicopathologic profile and treatment outcomes from a global multicenter registry. J Clin Oncol 2019;37:9081. [Crossref]
- Shin DH, Jo JY, Han JY. Dual Targeting of ERBB2/ERBB3 for the Treatment of SLC3A2-NRG1-Mediated Lung Cancer. Mol Cancer Ther 2018;17:2024-33. [Crossref] [PubMed]
- Jones MR, Lim H, Shen Y, et al. Successful targeting of the NRG1 pathway indicates novel treatment strategy for metastatic cancer. Ann Oncol 2017;28:3092-7. [Crossref] [PubMed]
- Cheema PK, Doherty M, Tsao MS. A Case of Invasive Mucinous Pulmonary Adenocarcinoma with a CD74-NRG1 Fusion Protein Targeted with Afatinib. J Thorac Oncol 2017;12:e200-2. [Crossref] [PubMed]
- Liu SV, Duruisseaux M, Tolba K, et al. 1969PTargeting NRG1-fusions in multiple tumour types: Afatinib as a novel potential treatment option. Ann Oncol 2019;30:mdz268.096.
- Kim HS, Han JY, Shin DH, et al. EGFR and HER3 signaling blockade in invasive mucinous lung adenocarcinoma harboring an NRG1 fusion. Lung Cancer 2018;124:71-5. [Crossref] [PubMed]
- Sequist LV, Yang JCH, Yamamoto N, et al. Phase III study of afatinib or cisplatin plus pemetrexed in patients with metastatic lung adenocarcinoma with EGFR mutations. J Clin Oncol 2013;31:3327-34. [Crossref] [PubMed]
- Wu YL, Zhou C, Hu CP, et al. Afatinib versus cisplatin plus gemcitabine for first-line treatment of Asian patients with advanced non-small-cell lung cancer harbouring EGFR mutations (LUX-Lung 6): an open-label, randomised phase 3 trial. Lancet Oncol 2014;15:213-22. [Crossref] [PubMed]
- Soria JC, Felip E, Cobo M, et al. Afatinib versus erlotinib as second-line treatment of patients with advanced squamous cell carcinoma of the lung (LUX-Lung 8): an open-label randomised controlled phase 3 trial. Lancet Oncol 2015;16:897-907. [Crossref] [PubMed]
- Geuijen CAW, De Nardis C, Maussang D, et al. Unbiased Combinatorial Screening Identifies a Bispecific IgG1 that Potently Inhibits HER3 Signaling via HER2-Guided Ligand Blockade. Cancer Cell 2018;33:922-936.e10. [Crossref] [PubMed]
- Schram AM, Drilon A, Mercade TM, et al. 685TiPA phase II basket study of MCLA-128, a bispecific antibody targeting the HER3 pathway, in NRG1 fusion-positive advanced solid tumours. Ann Oncol 2019;30:mdz247.169.
- Passiglia F, Caparica R, Giovannetti E, et al. The potential of neurotrophic tyrosine kinase (NTRK) inhibitors for treating lung cancer. Expert Opin Investig Drugs 2016;25:385-92. [Crossref] [PubMed]
- Vaishnavi A, Capelletti M, Le AT, et al. Oncogenic and drug-sensitive NTRK1 rearrangements in lung cancer. Nat Med 2013;19:1469-72. [Crossref] [PubMed]
- Cocco E, Scaltriti M, Drilon A. NTRK fusion-positive cancers and TRK inhibitor therapy. Nat Rev Clin Oncol 2018;15:731-47. [Crossref] [PubMed]
- Amatu A, Sartore-Bianchi A, Siena S. NTRK gene fusions as novel targets of cancer therapy across multiple tumour types. ESMO Open 2016;1:e000023. [Crossref] [PubMed]
- Farago AF, Taylor MS, Doebele RC, et al. Clinicopathologic Features of Non-Small-Cell Lung Cancer Harboring an NTRK Gene Fusion. JCO Precis Oncol 2018;2018.
- Stransky N, Cerami E, Schalm S, et al. The landscape of kinase fusions in cancer. Nat Commun 2014;5:4846. [Crossref] [PubMed]
- Gatalica Z, Xiu J, Swensen J, et al. Molecular characterization of cancers with NTRK gene fusions. Mod Pathol 2019;32:147-53. [Crossref] [PubMed]
- Ou SHI, Sokol ES, Trabucco SE, et al. 1549PNTRK1-3 genomic fusions in non-small cell lung cancer (NSCLC) determined by comprehensive genomic profiling. Ann Oncol 2019;30:mdz260.071.
- Xia H, Xue X, Ding H, et al. Evidence of NTRK1 fusions as resistance mechanism to EGFR TKI in EGFR+ NSCLC. Results from a large-scale survey of NTRK1 fusions in Chinese lung cancer patients. Clin Lung Cancer 2019; [Epub ahead of print]. [PubMed]
- Russo A, Franchina T, Ricciardi GRR, et al. Central nervous system involvement in ALK-rearranged NSCLC: promising strategies to overcome crizotinib resistance. Expert Rev Anticancer Ther 2016;16:615-23. [Crossref] [PubMed]
- Bergethon K, Shaw AT, Ou SHI, et al. ROS1 rearrangements define a unique molecular class of lung cancers. J Clin Oncol 2012;30:863-70. [Crossref] [PubMed]
- Wilson TR, Sokol ES, Trabucco SE, et al. 443PDGenomic characteristics and predicted ancestry of NTRK1/2/3 and ROS1 fusion-positive tumours from >165,000 pan-solid tumours. Ann Oncol 2019;30:mdz244.005.
- Skoulidis F, Goldberg ME, Greenawalt DM, et al. STK11/LKB1 Mutations and PD-1 Inhibitor Resistance in KRAS-Mutant Lung Adenocarcinoma. Cancer Discov 2018;8:822-35. [Crossref] [PubMed]
- Remon J, Hendriks LE, Cabrera C, et al. Immunotherapy for oncogenic-driven advanced non-small cell lung cancers: Is the time ripe for a change? Cancer Treat Rev 2018;71:47-58. [Crossref] [PubMed]
- Solomon JP, Hechtman JF. Detection of NTRK Fusions: Merits and Limitations of Current Diagnostic Platforms. Cancer Res 2019;79:3163-8. [Crossref] [PubMed]
- Solomon JP, Linkov I, Rosado A, et al. NTRK fusion detection across multiple assays and 33,997 cases: diagnostic implications and pitfalls. Mod Pathol 2020;33:38-46. [Crossref] [PubMed]
- Penault-Llorca F, Rudzinski ER, Sepulveda AR. Testing algorithm for identification of patients with TRK fusion cancer. J Clin Pathol 2019;72:460-7. [Crossref] [PubMed]
- Marchiò C, Scaltriti M, Ladanyi M, et al. ESMO recommendations on the standard methods to detect NTRK fusions in daily practice and clinical research. Ann Oncol 2019;30:1417-27. [Crossref] [PubMed]
- Laetsch TW, Hawkins DS. Larotrectinib for the treatment of TRK fusion solid tumors. Expert Rev Anticancer Ther 2019;19:1-10. [Crossref] [PubMed]
- Hyman DM, van Tilburg CM, Albert CM, et al. 445PDDurability of response with larotrectinib in adult and pediatric patients with TRK fusion cancer. Ann Oncol 2019;30:mdz244.007.
- Rosen EY, Schram AM, Young RJ, et al. Larotrectinib Demonstrates CNS Efficacy in TRK Fusion-Positive Solid Tumors. JCO Precision Oncology 2019;1-5. [Crossref]
- Drilon A, Nagasubramanian R, Blake JF, et al. A Next-Generation TRK Kinase Inhibitor Overcomes Acquired Resistance to Prior TRK Kinase Inhibition in Patients with TRK Fusion-Positive Solid Tumors. Cancer Discov 2017;7:963-72. [Crossref] [PubMed]
- Hyman D, Kummar S, Farago A, et al. Abstract CT127: Phase I and expanded access experience of LOXO-195 (BAY 2731954), a selective next-generation TRK inhibitor (TRKi). Cancer Res 2019;79:CT127.
- Rolfo C, Ruiz R, Giovannetti E, et al. Entrectinib: a potent new TRK, ROS1, and ALK inhibitor. Expert Opin Investig Drugs 2015;24:1493-500. [Crossref] [PubMed]
- Drilon A, Siena S, Ou SHI, et al. Safety and Antitumor Activity of the Multitargeted Pan-TRK, ROS1, and ALK Inhibitor Entrectinib: Combined Results from Two Phase I Trials (ALKA-372-001 and. Cancer Discov 2017;7:400-9. [Crossref] [PubMed]
- De Braud FG, Siena S, Barlesi F, et al. 1488PDEntrectinib in locally advanced/metastatic ROS1 and NTRK fusion-positive non-small cell lung cancer (NSCLC): Updated integrated analysis of STARTRK-2, STARTRK-1 and ALKA-372-001. Ann Oncol 2019;30:mdz260.010.
- Rolfo C, Dziadziuszko R, Doebele RC, et al. 476PUpdated efficacy and safety of entrectinib in patients with NTRK fusion-positive tumors: Integrated analysis of STARTRK-2, STARTRK-1 and ALKA-372-001. Ann Oncol 2019;30:mdz244.038.
- Doebele RC, Dziadziuszko R, Drilon A, et al. LBA28Genomic landscape of entrectinib resistance from ctDNA analysis in STARTRK-2. Ann Oncol 2019;30:mdz394.017.
- Cocco E, Schram AM, Kulick A, et al. Resistance to TRK inhibition mediated by convergent MAPK pathway activation. Nat Med 2019;25:1422-7. [Crossref] [PubMed]
- Drilon A, Ou SHI, Cho BC, et al. Repotrectinib (TPX-0005) Is a Next-Generation ROS1/TRK/ALK Inhibitor That Potently Inhibits ROS1/TRK/ALK Solvent- Front Mutations. Cancer Discov 2018;8:1227-36. [Crossref] [PubMed]
- Drilon A, Cho BC, Kim DW, et al. 444PDSafety and preliminary clinical activity of repotrectinib in patients with advanced ROS1/TRK fusion-positive solid tumors (TRIDENT-1 study). Ann Oncol 2019;30:mdz244.006.
- Katayama R, Gong B, Togashi N, et al. The new-generation selective ROS1/NTRK inhibitor DS-6051b overcomes crizotinib resistant ROS1-G2032R mutation in preclinical models. Nat Commun 2019;10:3604. [Crossref] [PubMed]
- Chong CR, Bahcall M, Capelletti M, et al. Identification of Existing Drugs That Effectively Target NTRK1 and ROS1 Rearrangements in Lung Cancer. Clin Cancer Res 2017;23:204-13. [Crossref] [PubMed]
- Lin CC, Arkenau HT, Lu S, et al. A phase 1, open-label, dose-escalation trial of oral TSR-011 in patients with advanced solid tumours and lymphomas. Br J Cancer 2019;121:131-8. [Crossref] [PubMed]
- He AR, Cohen RB, Denlinger CS, et al. First-in-Human Phase I Study of Merestinib, an Oral Multikinase Inhibitor, in Patients with Advanced Cancer. Oncologist 2019;24:e930-42. [Crossref] [PubMed]
- Olmez I, Zhang Y, Manigat L, et al. Combined c-Met/Trk Inhibition Overcomes Resistance to CDK4/6 Inhibitors in Glioblastoma. Cancer Res 2018;78:4360-9. [Crossref] [PubMed]
Cite this article as: Russo A, Lopes AR, Scilla K, Mehra R, Adamo V, Oliveira J, Liu SV, Rolfo C. NTRK and NRG1 gene fusions in advanced non-small cell lung cancer (NSCLC). Precis Cancer Med 2020;3:14.

