Exploiting homologous recombination repair alterations in metastatic castrate-resistant prostate cancer: a case report and literature review
Introduction
An elderly male patient with heavily pretreated metastatic castrate-resistant prostate cancer (mCRPC) was found to have a germline PALB2 alteration along with other somatic alterations in HRR pathway genes and had a significant response to platinum followed by PARP inhibitor therapy.
Case presentation
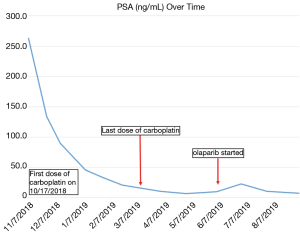
Our patient is a 70-year-old African-American male with a history of asthma, hyperlipidemia, and a spindle cell neoplasm of his mesentery treated with right hemicolectomy at age 52 who was diagnosed at age 60 with localized prostate cancer. Family history was notable for a mother diagnosed with breast cancer in her 60’s and a grandchild diagnosed with brain cancer at 18 months. His pre-treatment PSA was 7.5 ng/mL and a metastatic workup was negative. He underwent radical prostatectomy which showed a pathologic stage T3b/N0 adenocarcinoma with a Gleason score of 4+4 (tertiary pattern 5). He then completed a course of adjuvant radiation for positive surgical margins. He experienced a biochemical relapse over the next year with a PSA of 5 that was treated with androgen deprivation therapy (ADT) consisting of leuprolide injections given every 3 months along with denosumab. After an initial response, his PSA steadily increased over the next three years (PSA 4.3, doubling time 1.7 months) and imaging revealed bone metastases. Given progression to mCRPC enzalutamide was added to ADT followed by another prolonged biochemical response. Later his PSA began to increase and radium-223 was given without clear benefit. The PSA continued to rise and imaging showed progression of bone metastases and new pelvic and retroperitoneal lymphadenopathy. Subsequently he received 5 cycles of docetaxel and 3 cycles of cabazitaxel but again progressed radiographically (new liver and lung metastases), biochemically (peak PSA 2,628) and clinically with worsening fatigue, bone pain, weight loss, and bulky adenopathy-related leg edema. See Figure 1 for a timeline of significant events.
Tumor management
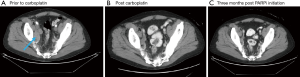
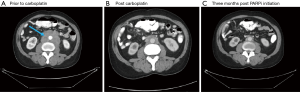
Next generation sequencing (NGS) was ordered from a prior biopsy specimen and revealed a microsatellite stable tumor with a tumor mutational burden of 8 mutations/Mb (intermediate by FoundOne), loss of FAS and PTEN, and mutations in CDK12, FANCA, KEL, and PALB2 [c.2878delC(p.Leu960Tyrfs*2)] (see Table S1). After consultation with the NGS service (Foundation One) the PALB2 mutation was deemed likely germline heterozygous and the patient was referred to Genetic Counseling. There an Integrated BRCA Analysis with Myriad myRisk Genetic testing was ordered and confirmed the heterozygous PALB2 mutation along with three variants of undetermined significance in ATM, BRIP1 and PMS2 (see Table S2). The patient was discussed at Molecular Tumor Board and the decision was made to proceed with carboplatin (AUC 6, every 3 weeks) due to initial cost issues with PARP inhibitors (PAPRi). He eventually received 7 doses with improvement in clinical symptoms and a decrease in his PSA from 2,628 to 62 (see Figure 2). Imaging at 4 months showed a mixed response then at 6 months marked improvement of osseous lesions on nuclear bone scan with decrease in size of retroperitoneal adenopathy and pulmonary nodules on CT (see Figures 3-5). Carboplatin was eventually discontinued due to fatigue and cytopenias. During this break from carboplatin PSA increased to 95. The PARPi olaparib was obtained with the help of a pharmaceutical company prescription savings program (AstaZeneca, AZ&ME). The patient tolerated olaparib 300 mg BID well and while at one month his PSA had risen from 95 to 224 by month two it had returned to 99 followed by a further decline to 57 at three months. Imaging at three months post PARPi initiation revealed a partial response with stable pulmonary and mediastinal nodules and further decreased abdominal nodules and bone lesions. He continues on the medication as of this writing.
Case-related literature review
Homologous recombination involves the use of a DNA template, such as a sister chromatid or homologous chromosome, to exchange nucleotide bases between DNA molecules. This process contributes to genetic variation in meiosis and is crucial in the repair of damaged DNA. Eukaryotic DNA is constantly undergoing damage, with as many as 105 DNA lesions of various kinds per day occurring in one cell (1). DNA damage repair (DDR) encompasses a variety of cellular mechanisms that, when defective, can lead to developmental issues, tumorigenesis, and cellular death.
Homologous recombination repair (HRR) has been called a conservative process in that it can recapitulate the original genetic sequence with high fidelity as opposed to some other process of DDR such as single strand annealing (SSA) or non-homologous end joining (NHEJ) (2). HHR is a means for cells to restore their genomic integrity in the setting of double stranded breaks (DSBs) but is also involved in repair of inter-strand crosslinks (3). Inter-strand crosslinking is one of the mechanisms via which alkylating agents, such as the nitrogen mustards, and platinum agents affect cellular toxicity.
DSBs are significantly toxic to cells and entail concurrent fractures of the phosphate backbones of complimentary DNA strands (4). These breaks can be caused by ionizing radiation, chemical or drug effects (i.e., bleomycin), through cell intrinsic processes such as when DNA polymerase encounters nicks in DNA or other means of stress related to genomic replication (5).
Recognition of genetic damage is the first step in DDR and the activation of oxidative-stress related proteins such as SIRT6 allow for recruitment of PARP1 to DSBs (6). This is followed rapidly (within seconds) by PARP1 regulated chromatin remodeling to allow for access to the affected strands of DNA (7). ATM is auto-phosphorylated in the setting of DBSs and, once activated, affects multiple downstream molecules including BRCA1 and histone variant H2AX, contributing to de-condensation of chromatin (1,8).
In HRR the MRN complex (nuclease Mre11-Rad50-Xrs2 with cofactor Sae2) binds to each broken end of DNA (9). The 5’ ends are resected to create 3’ overhangs of the complimentary DNA strands. Further 5 to 3 prime resections occur followed by binding of RPA to each 3’ overhanging single strand. With the help of mediators RAD51 recombinase overtakes the RPA binding and produces RAD51 filaments in an ATP dependent fashion that act as scaffolds (10). These mediators involve both positive regulators such as other RAD proteins (i.e., RAD52), BRCA1 and 2, and PALB2 and negative regulators such as FANCJ (9). These nucleoprotein filaments (often referred to as the presynaptic complex) then search for and locate a complimentary sequence of DNA scaffolds (10). Strand invasion occurs as the free 3’ filament interposes itself within the selected double stranded DNA thus displacing its homolog and forming base pairs with its complementary strand. This is known as the displacement loop or D-loop as it involves breaking of the previously connecting double stranded DNA base pairs (9). With the help of other proteins DNA polymerases are then able to extend the 3’ prime invading strand with the use of this complimentary sequence. The strand invasion process, D-loop extension, and eventual D-loop dissociation are likely influenced by many factors including the topography of the potential DNA partner strand, with a preference for negatively super-coiled domains (9).
Subsequently two primary models for completion of the DSB repair process exist, double strand break repair (DSBR) and synthesis-dependent strand annealing (SDSA). Briefly SDSA involves release of the extended invading strand, annealing of this strand to its original strand’s broken end, removal of excess DNA, further nucleotide synthesis to fill gaps followed by ligation without producing any crossover events (11). DSBR involves the creation of double Holiday junctions involving both the invading strand and the original 3’ DNA non-invading strand with the selected DS homologues. This is followed by further DNA synthesis and ligation, which, depending on the sites of cutting and homing of the recombination products, will most commonly lead to chromosomal crossover (11). In human cell DDR there is an apparent preference for SDSA, which is unsurprising given it does not lead to crossover events thus maintaining original genetic integrity (12).
There are different pathways by which DSBs can be repaired and competition between these processes exists intracellularly. Early binding of the NHEJ complex prevents HRR initialization due HRR’s requirement for a resected DNA end (2). The timing of the cell cycle also contributes to the type of DDR mechanism that cells utilize. HHR occurs once mammalian cells have produced sister chromatids so that these can be used as templates, thus it appears to be primarily active in the later S phase and G2 phase of the cell cycle when these chromatids are available. This temporal regulation may occur via decreased CDK meditated phosphorylation of BRCA2’s RAD51 binding site (13). Other repair processes, specifically the afore-mentioned NHEJ, predominate during other phases of the cell cycle and, given their mechanistic flexibility and template independence (14), are the primary means of DDR when cellular HHR is deficient. HHR may be more dynamic however, with some evidence that it may be able to overcome cell cycle restrictions (13).
Relevant to our patient PALB2 (Partner And Localizer of BRCA2) appears have multiple functions important to HRR including recruitment of RAD51 and BRCA2 to DSBs (15), binding BRCA2 and promoting the activity of the complex to promote the polymerization of the RAD51 filament (16), and assisting RAD51 polymers with strand invasion (17). The mutation found in our patient [c.2878delC(p.Leu960Tyrfs*2)] appears to result in a frameshift variant and the location of the deletion (WD40 domain) is likely involved in the binding of PALB2 to BRCA2, RAD51, and RAD51C thus affecting HRR (17,18). The finding that RAD51 is often up-regulated in BRCA1 deficient breast tumors suggests that the molecules in this process may compensate for each other in some malignancies (19). HRR defects promote tumorigenesis by forcing cells to rely on non-conservative and error-prone DDR mechanisms thus increasing mutational rate potentially leading to alterations that promote growth and cell survival. However the resultant malignant cells must balance the benefits of this increased rate of mutation with the potential for cellular death that can result from these errors.
Poly ADP-ribose polymerases (PARPs) compromise a group of enzymes that can catalyze the synthesis of PAR chains. The PARP enzymes (best characterized are PARP1 and PARP2) are relevant to DDR through their ability to recognize damage in DNA, bind to strand breaks and recruit enzymes that continue the DDR process. Their involvement in DDR was recognized after PARP levels were found to be increased after radiation damage to DNA and in the setting of increased DNA lesions in general (20). PARPs are involved in the processes of base excision repair and nucleotide excision repair in response to single stranded breaks (SSBs) in DNA (20). While SSBs in DNA are the thought to be the main target of PARPs, PARP1, as described earlier, is also implicated in the early stages DSB repair. Following DNA repair it is also necessary for the PARP complex to leave the site of DNA damage and there is evidence that PARP inhibitors may cause further cellular toxicity by inhibiting this post-repair dissociation thus stalling the replication fork (21). Synthetic lethality is the concept that while one defective process may not be enough to compromise cellular survival the additional inhibition of another related process may induce death. Thus the targeting of SSB repair (which if un-repaired can cause DSBs) and DDR sensing/initiation by PARP inhibition in the setting of deficient HHR may overwhelm the ability of tumor cells to survive (21).
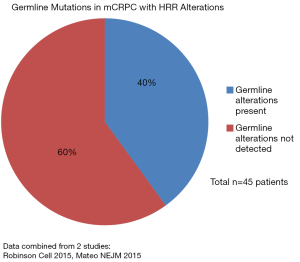
The frequency of DDR and more specifically HRR alterations have varied across studies. An analysis of the 333 primary prostate carcinoma samples revealed inactivation of DNA repair genes in 19% (22) with a more recent investigation showing an association between pathogenic DDR mutations in tumors and more advanced tumor stages and Gleason grade groups.(23) An examination of men with metastatic prostate cancer, unselected for family history or age, found germline mutations in DDR processes of 11.8% and that they were associated with worse prognoses (24,25). The most frequent germline mutations were in BRCA2 (5.35%), CHEK2 (1.87%) and ATM (1.59%) with BRCA1 (0.87%), RAD51D (0.43%) and PALB2 (0.43%) seen less often. Another study focusing on HRR in advanced prostate cancer included a subset of 221 patients with germline testing; 19% harbored a known or likely pathogenic germline mutation in an HRR gene. Of these 221 patients 27% were found to have any alteration (germline and/or somatic) in BRCA2, BRCA1, ATM or CHEK2 with germline analysis capturing only about half of those with likely HHR deficiency. PALB2 and ATM specifically were enriched in metastatic castrate-resistant versus metastatic castrate sensitive disease (26). For comparison in an Ashkenazi Jewish population of men with localized prostate cancer BRCA1 & 2 mutations were found in 3% (27). Another important point is that loss of function alterations in the HRR pathway often affects multiple alleles. Robinson et al reported biallelic BRCA2 loss in 90% of patients with BRCA2 alterations usually resulting from a somatic point mutation combined with loss of heterozygosity (28). The NCCN currently recommends germline evaluation of HHR genes (BRCA1, BRCA2, ATM, PALB2, CHEK2) and those involved in Lynch Syndrome/mismatch repair (MHL1, MSH2, MSH6, PMS2) for patients with high risk, very high risk, regionally advanced (N1) or metastatic prostate cancer; it states molecular testing of tumor tissue for these alterations should be considered (NCCN Guidelines Version 4.2019). See Figures 6 and 7 for HRR alteration frequency in data combined from multiple studies in mCRPC.

Our patient’s tumor sample also harbored mutations in CDK12 and FANCA both of which are involved in the process of DNA damage repair. A patient with metastatic prostate cancer with a FANCA germline mutation and somatic loss of heterozygosity of FANCA was reported to have a dramatic long term response to cisplatin-docetaxel with further in vitro studies confirming this mutation’s increased sensitivity to DNA damaging agents (29). Whether our patient’s sensitivity to platinum and PARP-inhibitor therapy was due to the germline PALB2 mutation combined with LOH versus the mutations in CDK12 and FANCA is unclear. The germline abnormalities of undetermined significance in ATM, BRIP1 and PSM2, which were not found in his tumor sample and have DDR functions, are also of interest in light of his personal and family cancer history.
Prior studies in unselected patients with metastatic prostate cancer treated with platinum agents resulted in modest clinical benefit without an increase in overall survival (30,31). As knowledge of DDR and HRR pathways and their sensitives has grown populations with these abnormalities have received more focus. A retrospective analysis targeting patients with HRR deficiency treated with carboplatin/docetaxel revealed extremely encouraging results with PSA responses in 75% of patients with germline BRCA2 mutations compared to 17% in those without (32). High response rates (88%) with PARPi have also been seen in patients with refractory prostate cancer who harbor DDR defects and were seen in patients with both germline and somatic alterations (33). More recent studies add further support to this treatment strategy. The TOPARP-B phase II trial treated mCRPC patients, selected for DDR alterations, with two different dose of olaparib. Varying radiologic response rates were seen depending on the genetic lesion with the highest rates in the BRCA1/2 (80%) and PABL2 (57%) mutated subgroups (34). The phase III PROfound trial compared olaparib to enzalutamide or abiraterone in patients with mCRPC with HRR alterations with the one group containing only those with BRCA1/2 or ATM mutations and the other including alterations of any HRR gene. Significant improvements in 12 month radiographic progression-free survival were seen in both groups (35). See Table 1 for preliminary PARPi Phase II trial results and Table 2 for ongoing Phase III clinical trials.
Table 1
| PARP inhibitor | Name | Phase | Condition | Mutation selection | Patients | Primary outcome | Results | NCT |
|---|---|---|---|---|---|---|---|---|
| Olaparib | TOPARP-B | Phase II | mCRPC Post progression on one or more taxanes | Selected for DDR alterations (germline or somatic, mono- or bi-allelic) by tumor sequencing | 98 patients randomized, 92 treated and evaluable | Response rate (RR): RECIST 1.1 and/or PSA decline >50% and/or circulating tumor cell count (CTC) conversion to less than 5 after 4 weeks | Olaparib 400 mg BID: RR 54%; olaparib 300 mg BID: RR 37%; with median f/u 17.6 months (mo), overall median PFS of 5.4 mo | NCT01682772 |
| Olaparib (300 mg BID) + Abiraterone vs. Abiraterone + placebo | Phase II | mCRPC; post treatment with docetaxel and candidates for abiraterone | Unselected for DDR or HRR alterations | 142 patients randomized, 71 to each arm | Investigator-assessed radiographic progression-free survival (rPFS) | Olaparib + Abiraterone: rPFS 13.8 mo; abiraterone + placebo: rPFS 8.2 mo; hazard Ratio 0.65 | NCT01972217 | |
| Olaparib (300 mg BID) vs. Abiraterone or Enzalutimade (Physicians Choice or PC) | PROfound | Phase III | mCRPC; post progression on prior new hormonal agent therapy | HRR alterations centrally and prospectively collected (foundation medicine) | Cohort A: 245 patients w/BRCA1/2 or ATM alterations; Cohort B: 142 patients with other HRR alterations | rPFS by blinded independent central review | Cohort A: Olaparib rPFS 7.39 vs. 3.55 mo for PC; Cohort A + B: Olaparib rPFS 5.82 vs. 3.52 mo for PC | NCT02987543 |
| Niraparib (300 mg daily) | GALAHAD | Phase II | mCRPC; post progression on androgen signaling inhibitors and taxanes | Selected for biallelic DNA-repair gene defects (DRD) by plasma assay | 123 patients with DRD, 39 with bi-allelic DRD mutations | Composite response rate (RR): RECIST 1.1, CTC conversion to less than 5/7.5 mL, PSA decline >50% | RR 65% in biallelic BRCA1/2 (n=23); RR 31% in biallelic non-BRCA (n=16) | NCT02854436 |
| Rucaparib (600 mg BID) | TRITON-2 | Phase II | mCRPC; post progression on 1–2 lines of androgen receptor therapy and 1 taxane | Germline or somatic alterations in BRCA1/2 or 13 other HRR genes | 52 patients treated | Centrally assessed objective response rate per modified RECIST v1.1/PCWG3 in patients with measurable disease and PSA decrease >50% in those without | Median f/u 3.7 mo; 11 of 23 (48%) evaluable BRCA altered patients had PSA response; 5 of 11 (46%) evaluable BRCA altered patients had radiographic response | NCT02952534 |
mCRPC, metastatic castrate-resistant prostate cancer.
Table 2
| Name | PARP inhibitor | Comparator | Condition | Primary outcome | NCT |
|---|---|---|---|---|---|
| PROpel | Olaparib + Abiraterone | Placebo + Abiraterone | First line in mCRPC | Radiologic Progression Free Survival | NCT0373280 |
| KEYLINK-010 | Olaparib + Pembrolizumab with Abiraterone or Enzalutamide | Abiraterone or Enzalutamide | mCRPC, unselected for HRR defects Post progression on one next gen hormonal agent | Overall Survival | NCT03834519 |
| – | Niraparib + Abiraterone | Abiraterone + Placebo | Metastatic Prostate Cancer, DDR alterations assessed pre-treatment | Radiographic Progression Free Survival | NCT03748641 |
| – | Talazoparib + Enzalutamide | Enzalutamide + Placebo | mCRPC, first line treatment once documented metastatic | Radiographic Progression Free Survival | NCT03395197 |
| TRITON3 | Rucaparib | Physician’s Choice: Abiraterone, Enzalutamide, or Docetaxel |
mCRPC, selected for HRR alterations | Radiographic Progression Free Survival | NCT02975934 |
mCRPC, metastatic castrate-resistant prostate cancer.
Conclusions
Once a defect is detected in the HHR pathway there is a clear rationale for use of inter-strand cross-linking agents or PARP inhibition as well as other potential methods that, via DNA damage, would force cells to rely on more error prone repair mechanisms. Responses appear to occur regardless of whether the alterations in the HHR pathway are germline or somatic thus lending support for the evaluation of both in patients with advanced disease. In metastatic CRPC refractory to standard treatments a combination of germline and somatic testing seems a reasonable clinical algorithm based on mounting evidence for actionable mutations in this disease.
Future areas to explore include new targets for simultaneous inhibition of different DDR mechanisms. For example would concurrently targeting topoisomerase be useful as it is likely necessary for the strand invasion process in HHR or would it be limited by excessive toxicity (36,37)? What are the ways in which tumors harboring DDR defects evolve to balance the positive and negative selection effects of mutagenesis from increased error rate? Recent evidence has shown improved radiographic progression free survival in patients with mCRPC treated with olaparib and abiraterone who were not selected for HRR alterations (38). In the potential setting of general benefit from PARP inhibition combinations in mCRPC, will HRR alterations select for better or longer responses? How can the relationships between androgen receptor signaling, the PI3K-AKT pathway and HRR deficiencies be further exploited with therapy (39)? Beyond specific mutations will BRCAness or BRCA-like genetic scars also correlate with response to platinum agents and PARPi as seen in ovarian cancer? Lastly the ability of malignancies to adapt to these therapies is becoming more apparent and offers an area ripe for further inquiry (40).
Table S1
| Microsatellite status | MS-Stable |
|---|---|
| Tumor mutational burden | 8 Mut/Mb |
| CDK12 | L836fs*16 |
| FANCA | S251* |
| FAS | Loss |
| KEL | Splice site 348_400+158del211 |
| PALB2 | L960fs*2 |
| PTEN | Loss |
Table S2
| Gene | Mutation | Interpretation |
|---|---|---|
| PALB2 | c.2878delC (p.Leu960Tyrfs*2) Heterozygous | High cancer risk |
| ATM | c.290T>C (p.Ile97Thr) | Uncertain clinical significance |
| BRIP1 | c.1057>C (p.Tyr353His) | Uncertain clinical significance |
| PMS2 | c.2179C>g (p.Gln727Glu) (aka Q727E (2179C>G)) | Uncertain clinical significance |
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editor Balazs Halmos for the series “Precision Oncology Tumor Board” published in Precision Cancer Medicine. The article has undergone external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/pcm.2020.01.02). The Series “Precision Oncology Tumor Board” was commissioned by the editorial office without any funding or sponsorship. BG reports personal fees from Janssen, Eisai, and Genomic Health, outside the submitted work. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and national research committees and with the Helsinki Declaration (as revised in 2013). Written informed consent was obtained from the patient for publication of this manuscript and any accompanying images.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Liu B, Yip RK, Zhou Z. Chromatin remodeling, DNA damage repair and aging. Curr Genomics 2012;13:533-47. [Crossref] [PubMed]
- Moynahan ME, Jasin M. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat Rev Mol Cell Biol 2010;11:196-207. [Crossref] [PubMed]
- Huang Y, Li L. DNA crosslinking damage and cancer - a tale of friend and foe. Transl Cancer Res 2013;2:144-54. [PubMed]
- Mehta A, Haber JE. Sources of DNA double-strand breaks and models of recombinational DNA repair. Cold Spring Harb Perspect Biol 2014;6:a016428. [Crossref] [PubMed]
- Goodsell DS. The Molecular Perspective: Double Stranded DNA Breaks. Oncologist 2005;361-2. [Crossref] [PubMed]
- Van Meter M, Simon M, Tombline G, et al. JNK Phosphorylates SIRT6 to Stimulate DNA Double-Strand Break Repair in Response to Oxidative Stress by Recruiting PARP1 to DNA Breaks. Cell Rep 2016;16:2641-50. [Crossref] [PubMed]
- Sellou H, Lebeaupin T, Chapuis C, et al. The poly(ADP-ribose)-dependent chromatin remodeler Alc1 induces local chromatin relaxation upon DNA damage. Mol Biol Cell 2016;27:3791-9. [Crossref] [PubMed]
- Rogakou EP, Pilch DR, Orr AH, et al. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem 1998;273:5858-68. [Crossref] [PubMed]
- Wright WD, Shah SS, Heyer WD. Homologous recombination and the repair of DNA double-strand breaks. J Biol Chem 2018;293:10524-35. [Crossref] [PubMed]
- Kowalczykowski SC. An Overview of the Molecular Mechanisms of Recombinational DNA Repair. Cold Spring Harb Perspect Biol 2015; [Crossref] [PubMed]
- Sung P, Kelin H. Mechanism of homologous recombination: mediators and helicases take on regulatory functions. Nat Rev Mol Cell Biol 2006;739-50. [Crossref] [PubMed]
- Zapotoczny G, Sekelsky J. Human Cell Assays for Synthesis-Dependent Strand Annealing and Crossing over During Double-Strand Break Repair. G3 (Bethesda) 2017;7:1191-9. [Crossref] [PubMed]
- Esashi F, Christ N, Gannon J, et al. CDK-dependent phosphorylation of BRCA2 as a regulatory mechanism for recombinational repair. Nature 2005;434:598-604. [Crossref] [PubMed]
- Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem 2010;79:181-211. [Crossref] [PubMed]
- Sy SM, Huen MS, Zhu Y, et al. PALB2 regulates recombinational repair through chromatin association and oligomerization. J Biol Chem 2009;18302-10. [Crossref] [PubMed]
- Al Abo M, Dejsuphong D, Hirota K, et al. Compensatory functions and interdependency of the DNA-binding domain of BRCA2 with the BRCA1-PALB2-BRCA2 complex. Cancer Res 2014;74:797-807. [Crossref] [PubMed]
- Buisson R, Niraj J, Pauty J, et al. Breast cancer proteins PALB2 and BRCA2 stimulate polymerase η in recombination-associated DNA synthesis at blocked replication forks. Cell Rep 2014;6:553-64. [Crossref] [PubMed]
- Park JY, Singh TR, Nassar N, et al. Breast cancer-associated missense mutants of the PALB2 WD40 domain, which directly binds RAD51C, RAD51 and BRCA2, disrupt DNA repair. Oncogene 2014;33:4803-12. [Crossref] [PubMed]
- Martin RW, Orelli BJ, Yamazoe M, et al. RAD51 up-regulation bypasses BRCA1 function and is a common feature of BRCA1-deficient breast tumors. Cancer Res 2007;67:9658-65. [Crossref] [PubMed]
- Morales J, Li L, Fattah FJ, et al. Review of poly (ADP-ribose) polymerase (PARP) mechanisms of action and rationale for targeting in cancer and other diseases. Crit Rev Eukaryot Gene Expr 2014;24:15-28. [Crossref] [PubMed]
- Lord CJ, Ashworth A. PARP inhibitors: Synthetic lethality in the clinic. Science 2017;355:1152-8. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. The Molecular Taxonomy of Primary Prostate Cancer. Cell 2015;163:1011-25. [Crossref] [PubMed]
- Marshall CH, Fu W, Wang H, et al. Prevalence of DNA repair gene mutations in localized prostate cancer according to clinical and pathologic features: association of Gleason score and tumor stage. Prostate Cancer Prostatic Dis 2019;22:59-65. [Crossref] [PubMed]
- Pritchard CC, Mateo J, Walsh MF, et al. Inherited DNA-Repair Gene Mutations in Men with Metastatic Prostate Cancer. N Engl J Med 2016;375:443-53. [Crossref] [PubMed]
- Castro E, Goh C, Olmos D, et al. Germline BRCA mutations are associated with higher risk of nodal involvement, distant metastasis, and poor survival outcomes in prostate cancer. J Clin Oncol 2013;31:1748-57. [Crossref] [PubMed]
- Abida W, Armenia J, Gopalan A, et al. Prospective Genomic Profiling of Prostate Cancer Across Disease States Reveals Germline and Somatic Alterations That May Affect Clinical Decision Making. JCO Precis Oncol 2017. doi:
10.1200/PO.17.00029 . - Gallagher DJ, Gaudet MM, Pal P, et al. Germline BRCA mutations denote a clinicopathologic subset of prostate cancer. Clin Cancer Res 2010;16:2115-21. [Crossref] [PubMed]
- Robinson D, Van Allen EM, Wu YM, et al. Integrative clinical genomics of advanced prostate cancer. Cell 2015;161:1215-28. [Crossref] [PubMed]
- Wilkes DC, Sailer V, Xue H, et al. A germline FANCA alteration that is associated with increased sensitivity to DNA damaging agents. Cold Spring Harb Mol Case Stud 2017; [Crossref] [PubMed]
- Sternberg CN, Petrylak DP, Sartor O, et al. Multinational, double-blind, phase III study of prednisone and either satraplatin or placebo in patients with castrate-refractory prostate cancer progressing after prior chemotherapy: the SPARC trial. J Clin Oncol 2009;27:5431-8. [Crossref] [PubMed]
- Ross RW, Beer TM, Jacobus S, et al. A phase 2 study of carboplatin plus docetaxel in men with metastatic hormone-refractory prostate cancer who are refractory to docetaxel. Cancer 2008;112:521-6. [Crossref] [PubMed]
- Pomerantz MM, Spisák S, Jia L, et al. The association between germline BRCA2 variants and sensitivity to platinum-based chemotherapy among men with metastatic prostate cancer. Cancer 2017;123:3532-9. [Crossref] [PubMed]
- Mateo J, Carreira S, Sandhu S, et al. DNA-Repair Defects and Olaparib in Metastatic Prostate Cancer. N Engl J Med 2015;373:1697-708. [Crossref] [PubMed]
- Mateo J, Porta N, Mcgovern UB, et al. TOPARP-B: A phase II randomized trial of the poly(ADP)-ribose polymerase (PARP) inhibitor olaparib for metastatic castration resistant prostate cancers (mCRPC) with DNA damage repair (DDR) alterations. J Clin Oncol 2019;37:abstr 5005.
- De Bono JS, Hussain M, Thiery-Vuillemin A, et al. PROfound: A randomized Phase III trial evaluating olaparib in patients with metastatic castration-resistant prostate cancer and a deleterious homologous recombination DNA repair aberration. Ann Oncol 2019;35:v851-v934.
- Xu Y, Her C. Inhibition of Topoisomerase (DNA) I (TOP1): DNA Damage Repair and Anticancer Therapy. Biomolecules 2015;5:1652-70. [Crossref] [PubMed]
- Nickoloff JA, Jones D, Lee SH, et al. Drugging the Cancers Addicted to DNA Repair. J Natl Cancer Inst 2017; [Crossref] [PubMed]
- Clarke N, Wiechno P, Alekseev B, et al. Olaparib combined with abiraterone in patients with metastatic castration-resistant prostate cancer: a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet Oncol 2018;19:975-86. [Crossref] [PubMed]
- Virtanen V, Paunu K, Ahlskog JK, et al. PARP Inhibitors in Prostate Cancer—The Preclinical Rationale and Current Clinical Development. Genes (Basel) 2019; [Crossref] [PubMed]
- Goodall J, Mateo J, Yuan W, et al. Circulating Cell-Free DNA to Guide Prostate Cancer Treatment with PARP Inhibition. Cancer Discov 2017;7:1006-17. [Crossref] [PubMed]
Cite this article as: Peeke SZ, Lopez R, Gartrell B. Exploiting homologous recombination repair alterations in metastatic castrate-resistant prostate cancer: a case report and literature review. Precis Cancer Med 2020;3:10.