Annotating the next generation sequencing report
Introduction
With the advent of precision oncology, a growing number of genetic alterations need to be tested in tumor biopsies, particularly in certain malignancies. In the case of advanced lung adenocarcinoma, the list of driver alterations that have been described is quite extensive, including mutations in KRAS, EGFR, HER2, BRAF or NRAS; splicing variants of MET exon 14 and fusions involving ALK, ROS1, RET, NTRK1 or NRG1 (1,2). In other types of solid or hematological malignancies, the number of driver genes is rarely less than 10 (3). In addition, non-driver mutations in genes such as TP53 or PIK3CA or copy number variations (CNVs) of MET, HER2, FGFR1 and other genes might also have clinical relevance (4,5). Finally, testing the expression of certain proteins such as PD-L1 or HER2 by immunohistochemistry is mandatory in many tumor types (6-9). In order to analyze this increasing number of genetic alterations, many clinical laboratories are incorporating multiplexed techniques. To this end, there are several strategies and technologies available, which can detect different types of alterations. Although DNA-based next generation sequencing techniques (NGS) are becoming increasingly popular, they cannot determine gene or protein expression levels and they have also been described to miss a certain number of gene fusions and splice variants (10). Consequently, oncology-centered clinical laboratories need to complement NGS with IHC or RNA-based techniques in order to comprehensively analyze tumor samples. In our laboratory, we currently test somatic and hereditary mutations and CNVs by NGS; gene fusions and splicing variants using nCounter, a multiplexed RNA-based platform; and PD-L1 expression by IHC. In the limited number of cases where non-conclusive results are obtained for CNVs or fusions, IHC or FISH are used for confirmatory purposes.
Panels in multiplexed platforms
A relevant decision that clinical laboratories and oncology departments need to face is the choice of a panel for NGS or other multiplexed platforms. In the case of NGS, several panels are commercially available, and customized panels can be easily designed and synthesized by specialized companies. The number of genes to be sequenced goes from 20 to more than 300 and has a direct impact in the cost of analysis, which is a key issue towards the full implementation of NGS in the routine clinical practice (11,12).
In order to select or customize a panel, a first question is the range of tumors that is going to be analyzed. In our laboratory, the choice was to design a custom panel that could be comprehensively used in all types of solid tumors. Then, the specific genes and exons within each gene targeted by the panel have to be selected. If the panel has to be clinically useful, it must be able to determine all genetic alterations detectable on DNA with an associated approved therapy, such as EGFR or BRAF mutations (see Table 1). In addition, it should include “emerging” biomarkers that have been demonstrated to predict response to a targeted therapy but have not yet received approval by the health authorities. Some examples are MET exon 14 splicing variants, MET amplification or HER2 mutations. Also, if the oncology department has clinical trials of new drugs targeting specific alterations, it might be convenient to include them in the panel in order to facilitate recruitment. Finally, it can also be useful to include genes that have been reported to have prognostic value or to be associated with acquired or intrinsic resistance to therapies. As an example, Table 2 presents the list of genes selected for the NGS custom panel we use in our institution. The panel is currently being employed for tissue and liquid biopsy samples of cancer patients, baseline and at progression (13-15).
Table 1
| Biomarker | Malignancies | Technique(s) |
|---|---|---|
| Expression of estrogen receptors (ER) | Breast cancer | IHC |
| Expression of progesterone receptors (PR) | Breast cancer | IHC |
| HER2 overexpression/amplification | Breast cancer | IHC/FISH |
| KRAS mutations | CRC | DNA-based |
| NRAS mutations | CRC | DNA-based |
| BRAF mutations | CRC, NSCLC | DNA-based |
| EGFR mutations | NSCLC | DNA-based |
| ALK fusions | NSCLC | FISH, IHC, DNA or RNA-based |
| ROS1 fusions | CRC, NSCLC | FISH, DNA or RNA-based |
| KIT mutations | GIST | DNA-based |
| PD-L1 expression | Several | IHC |
Table 2
| Gene | CNVs analysis | Exons covered |
|---|---|---|
| ALK | NO | 22, 23, 24, 25 |
| BRAF | YES | 11, 15 |
| CDK4 | YES | – |
| CDK6 | YES | – |
| EGFR | YES | 12, 18, 19, 20, 21 |
| ERBB2 | YES | 8, 17, 19, 20, 21 |
| ERBB4 | NO | 8, 17, 19, 20, 21 |
| FGFR1 | YES | – |
| IDH1 | NO | 4 |
| IDH2 | NO | 4 |
| KIT | NO | 8, 9, 11, 13, 14, 17 |
| KRAS | YES | 2, 3, 4 |
| MET | YES | 13, 15 (splicing MET14) |
| NRAS | NO | 2, 3, 4 |
| PDGFRA | NO | 12, 18 |
| PIK3CA | NO | 10, 21 |
| RICTOR | YES | – |
| ROS1 | NO | 38 |
| STK11 | NO | 2, 3, 4, 5, 6, 7, 8, 9 |
| TP53 | NO | 4, 5, 6, 7, 8, 9 |
Annotating variants—overview
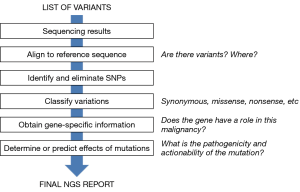
NGS analysis of a tumor sample usually identifies a large number of nucleotide changes relative to the reference human genome, these changes are usually referred to as “variants”. Even if the number of genes and exons included in the NGS panel is limited, the number of variants can be significant. Table 3 presents the results of a NGS analysis of two FFPE biopsies using the 20 gene panel designed at our institution. Typically, the number of variants detected after alignment goes from 15 to more than 50. When using larger panels, the number of variants increases accordingly. The challenge is then annotation, which is the process of identifying the functional and clinical relevance of the variants so that the final genomic report clearly identifies the actionable alterations that can predict sensitivity/resistance to therapies or alter prognosis (16). Annotation can be quite complex, and surveys have demonstrated that oncologists are not fully confident in their ability to interpret information derived from NGS analyses (17). Pathologists also lack the proper background, so the annotation process requires the participation of molecular biologists, very particularly in its first steps. Although many clinicians and health care institutions are extremely reluctant to incorporate molecular biologists in their staffs, circumscribing them to research activities; it is becoming increasingly clear that they must necessarily have an active role in molecular testing and annotation, granting them a place in the tumor boards already functioning in many hospitals (3). After all, in many tumor types, the information derived from NGS is at least as relevant to the selection of therapies as the information coming from imaging techniques or pathological assessments (18-21).
Table 3
| Gene | c. variant | p. variant | VAF | F Count | R Count | Reported |
|---|---|---|---|---|---|---|
| NRAS | c.406A>G | p.Ser136Gly | 7.04% | 0 | 10 | |
| IDH1 | c.347delA | p.Asn116fs | 4.20% | 0 | 5 | |
| ERBB4 | c.884-7delT | (Intronic) | 11.84% | 16 | 2 | |
| ERBB4 | c.884-8_884-7delTT | (Intronic) | 10.32% | 12 | 4 | |
| ERBB4 | c.884-9_884-7delTTT | (Intronic) | 4.52% | 5 | 2 | |
| PIK3CA | c.1540-6T>G | (Intronic) | 15.12% | 0 | 13 | |
| PDGFRA | c.1701A>G | (Intronic) | 99.26% | 50 | 85 | |
| PDGFRA | c.2472C>T | (Intronic) | 51.87% | 37 | 74 | |
| RICTOR | c.754-7_754-6insT | (Intronic) | 6.10% | 3 | 2 | |
| RICTOR | c.754-7delT | (Intronic) | 8.28% | 5 | 7 | |
| EGFR | c.2361G>A | (Intronic) | 47.78% | 23 | 20 | |
| MET | c.3109G>A | p.Gly1037Ser | 3.05% | 0 | 3 | |
| MET | c.3912C>T | (Intronic) | 6.00% | 5 | 1 | |
| MET | c.4071G>A | (Intronic) | 51.41% | 34 | 39 | |
| MET | c.4146G>A | (Intronic) | 39.86% | 7 | 52 | |
| BRAF | c.2128-5delT | (Intronic) | 49.43% | 41 | 46 | |
| KRAS | c.38G>A | p.Gly13Asp | 8.78% | 16 | 2 | KRAS p.Gly13Asp |
| CDK4 | c.662A>G | p.Asp221Gly | 35.59% | 39 | 24 | |
| TP53 | c.920-10T>C | (Intronic) | 5.88% | 2 | 2 | |
| TP53 | c.592G>T | p.Glu198* | 4.40% | 4 | 3 | TP53 p.Glu198* |
| TP53 | c.215C>G | p.Pro72Arg | 57.98% | 23 | 46 |
The variants reported after filtering and annotation are listed in the last column. VAF, variant alellic fraction. F, forward. R, revers.
Figure 1 summarizes the different steps of the annotation process. In each step, variants are filtered so that, at the end of the process, only a few clinically relevant variants should remain, that are sometimes accompanied in the final NGS report by a short number of variants of uncertain significance.
Alignment and first filtering based on frequencies and coverage
The first relevant issue that needs to be considered when annotating variants is the quality of the sequencing. The software programs associated with the NGS platforms generate a variety of quality parameters for the sequencing of each sample, such as coverage, GC percentage, ambiguous base content and other quality scores. Ideally, certain thresholds need to be met for a sequencing result to be considered reliable. In addition, all the gene regions of interest have to be adequately covered (22). For instance, in a non-small cell lung cancer (NSCLC) sample, it would not be acceptable that exon 19 of the EGFR is not sufficiently covered.
During data analysis, sequencing results of the regions targeted by the panel are first aligned to a reference sequence. There are currently software tools that automatically perform these steps and generate two types of computer files: SAM or BAM files, used to represent aligned sequences; and VAF files, which compile the information about the variants found at specific positions in the sample analyzed relative to a reference genome. Some programs allow for visualization of these files. In the case of VAF files, a list of variants is generated, accompanied with additional data such as allelic fraction, number of counts in forward and reverse, etc. When reporting variants, ambiguity needs to be avoiding meaning that the transcript and respective coding and protein position should be mentioned in the report. Ultimately, the genomic coordinate will also unambiguous characterize the variant. All this data should ideally be generated with the software used; if this is not the case, the person responsible for the report should produce it using adequate databases and programs (Table 4) (16,23,24).
Table 4
| Process | Tool/database | Website |
|---|---|---|
| Sequence alignment | Burrows-Wheeler Aligner | http://bio-bwa.sourceforge.net/ |
| T-Coffee | http://tcoffee.org/ | |
| Variant calling | GATK | https://software.broadinstitute.org/gatk/ |
| Free Bayes | https://github.com/ekg/freebayes | |
| Variant annotation (tools) | Annotate Variation (ANNOVAR) | http://annovar.openbioinformatics.org/en/latest/ |
| Polymorphism Phenotyping v2 (PolyPhen-2) | http://genetics.bwh.harvard.edu/pph2/ | |
| Sorting Intolerant from Tolerant (SIFT) | https://sift.bii.a-star.edu.sg/ | |
| Ensembl Variant Effect Predictor (VEP) | https://www.ensembl.org/info/docs/tools/vep/index.html | |
| Variant annotation (databases) | Catalogue of Somatic Mutations in Cancer (COSMIC) | https://cancer.sanger.ac.uk/cosmic |
| cBioPortal | http://www.cbioportal.org/ | |
| The Cancer Genome Atlas (TCGA) | https://portal.gdc.cancer.gov/ | |
| SNP database (dbSNP) | https://www.ncbi.nlm.nih.gov/snp/ | |
| Precision Oncology decision support | Personalized Cancer Therapy | https://pct.mdanderson.org/ |
| My Cancer Genome | https://www.mycancergenome.org/ | |
| Precision Oncology Knowledge Base (OncoKB) | https://oncokb.org/ |
A first analysis and filtering of variants can be performed based on the data contained in the list of variants and other quality parameters of the NGS run. Variants with very low counts only in forward or reverse are very likely to be artifacts, particularly in the case of FFPE samples. This might also be the case of variants with very low allelic fractions. For instance, a base replacement with a 0.4% allelic fraction in a tissue sample with 90% tumor infiltration is probably artifactual. Or, in case it was really present, it would just affect a very minor clone of tumor cells and, in all likelihood, will not have any clinical relevance. Many software tools that generate list of variants also incorporate some filters in order to eliminate such low abundance variants. In some cases, the thresholds of those filters can be re-set by the operator.
Identification of actionable variants—functional annotation
Actionable variants are those with potential clinical utility, either because they alter prognosis or because they predict sensitivity or resistance to specific therapies (25-27). In the clinical setting, a particular emphasis is placed on therapeutic actionability, since it can guide the treatment of the patient. This kind of actionability should not be restricted to EMA or FDA-approved therapies and markers, but should also include variants that allow the enrolment of patients into clinical trials (3).
In a significant percentage of tumor samples, well-known actionable mutations appear in the filtered list of variants. Most colon cancer samples present mutations in codons 12, 13 and 61 of the KRAS gene or 600 of the BRAF gene that predict resistance to anti-EGFR antibodies, while 15–50% of NSCLC samples harbor the p.L858R mutation or small insertions/deletions (indels) in the EGFR gene, which make the patient eligible for EGFR TKI therapies. These variants can be directly annotated, while the rest will have to go through the process of functional annotation.
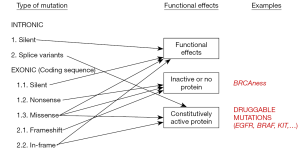
The first step in this process is to eliminate single nucleotide polymorphisms (SNPs), germ-line variants that are present in a significant percentage of the human population. In patient samples, SNPs appear at allelic frequencies close to 50%. Software programs are available in some NGS platforms that automatically identify SNPs but, if this is not the case, publicly available databases can be used (Table 4) (3,16). The next step is to determine the functional effects of the variation (Figure 2). A change in the nucleotide sequence can be silent or synonymous and have no effect on the amino-acid sequence of the corresponding protein. Obviously, these types of mutations are not expected to be actionable. Other mutations do introduce changes the amino-acid sequence, but their effects on the function of the protein depend on many factors. Frameshift deletions or nonsense mutations, which introduce a premature stop codon, are likely to generate an inactive protein, except if they are located at the very end of the gene. Small in-frame insertion-deletions (indels) located in the exons coding for the kinase domain of a membrane receptor are typically associated with constitutive activation of the protein. Finally, missense mutations, which replace a single amino-acid, will have a functional effect depending on several factors, such as the type of amino-acid replacement (conservative or not) and the localization of the replacement within the protein sequence (active center, ligand-binding regions, etc.) (Figure 2). The gene affected by the mutation is also relevant for functional annotation, particularly if it has been described to be an oncogene or a tumor suppressor.
A number of computer algorithms have been developed to predict the functional effect of mutations (Table 4), usually based on cross-species conservation but incorporating other considerations (28). However, the predictive power of these algorithms is limited, and their sensitivity and specificity have been estimated to be around 65–75% (29). In particular, they often fail to identify mutations leading to constitutive activation (30). Publicly accessible database are also useful in the process of functional annotation. PubMed, somatic mutation databases (COSMIC and TCGA) or precision oncology websites (Table 4) can be used to search for a specific variation, check if it has been previously identified in tumor samples and find out about its functional effects and actionability. At the end of this process of functional annotation, most variants should be adequately classified. However, a significant percentage of tumor samples harbor variants from which there is no functional information available and cannot be properly categorized. They are the so-called variants of uncertain significance (VUS).
The final NGS report
The process of filtering and annotating variants described below should dramatically reduce the initial pre-analysis list of variants. Table 3 presents the final list of variants in a tumor sample analyzed in our institution. The final NGS report to be handed to the clinician should clearly categorize the variants, listing first the therapeutic actionable variants and the approved drugs or clinical trials associated with them; followed by other variants with prognostic or predictive significance and finally the VUS, if any. In those cases where several actionable variants are found, they should be prioritized according to several parameters. Alterations with targeted agents approved by the health authorities and ideally covered by the public health care system or the insurance companies should go first. Then, variants with drugs with proven clinical efficacy not yet approved but that can be obtained from pharmaceutical companies through programs of compassionate use. Finally, alterations allowing enrolment of the patient in clinical trials, preferentially in the same hospital or hospitals nearby. If the clinical institution has a tumor board, the NGS report should be presented there, and the board should examine and discuss it in order to select the best treatment option for the patient (27,31,32).
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editor Nir Peled for the series dedicated to the Congress on Clinical Controversies in Lung Cancer (CCLC 2018) published in Precision Cancer Medicine. The article has undergone external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/pcm.2019.11.02). The series dedicated to the Congress on Clinical Controversies in Lung Cancer (CCLC 2018) was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bubendorf L, Lantuejoul S, de Langen AJ, et al. Non-small cell lung carcinoma: diagnostic difficulties in small biopsies and cytological specimens: Number 2 in the Series "Pathology for the clinician" Edited by Peter Dorfmuller and Alberto Cavazza. Eur Respir Rev 2017;26. doi:
10.1183/16000617.0007-2017 . - Kerr KM, Dafni U, Schulze K, et al. Prevalence and clinical association of gene mutations through multiplex mutation testing in patients with NSCLC: results from the ETOP Lungscape Project. Ann Oncol 2018;29:200-8. [Crossref] [PubMed]
- Kurnit KC, Dumbrava EEI, Litzenburger B, et al. Precision Oncology Decision Support: Current Approaches and Strategies for the Future. Clin Cancer Res 2018;24:2719-31. [Crossref] [PubMed]
- Molina-Vila MA, Bertran-Alamillo J, Gasco A, et al. Nondisruptive p53 mutations are associated with shorter survival in patients with advanced non-small cell lung cancer. Clin Cancer Res 2014;20:4647-59. [Crossref] [PubMed]
- Murtuza A, Bulbul A, Shen JP, et al. Novel Third-Generation EGFR Tyrosine Kinase Inhibitors and Strategies to Overcome Therapeutic Resistance in Lung Cancer. Cancer Res 2019;79:689-98. [Crossref] [PubMed]
- Hirsch FR, McElhinny A, Stanforth D, et al. PD-L1 Immunohistochemistry Assays for Lung Cancer: Results from Phase 1 of the Blueprint PD-L1 IHC Assay Comparison Project. J Thorac Oncol 2017;12:208-22. [Crossref] [PubMed]
- Thunnissen E, de Langen AJ, Smit EF. PD-L1 IHC in NSCLC with a global and methodological perspective. Lung Cancer 2017;113:102-5. [Crossref] [PubMed]
- Udall M, Rizzo M, Kenny J, et al. PD-L1 diagnostic tests: a systematic literature review of scoring algorithms and test-validation metrics. Diagn Pathol 2018;13:12. [Crossref] [PubMed]
- Wesola M, Jelen M. A Comparison of IHC and FISH Cytogenetic Methods in the Evaluation of HER2 Status in Breast Cancer. Adv Clin Exp Med 2015;24:899-903. [Crossref] [PubMed]
- Benayed R, Offin M, Mullaney K, et al. High Yield of RNA Sequencing for Targetable Kinase Fusions in Lung Adenocarcinomas with No Mitogenic Driver Alteration Detected by DNA Sequencing and Low Tumor Mutation Burden. Clin Cancer Res 2019;25:4712-22. [Crossref] [PubMed]
- van Nimwegen KJ, van Soest RA, Veltman JA, et al. Is the $1000 Genome as Near as We Think? A Cost Analysis of Next-Generation Sequencing. Clin Chem 2016;62:1458-64. [Crossref] [PubMed]
- Trosman JR, Weldon CB, Kelley RK, et al. Challenges of coverage policy development for next-generation tumor sequencing panels: experts and payers weigh in. J Natl Compr Canc Netw 2015;13:311-8. [Crossref] [PubMed]
- Mayo-de-Las-Casas C, Garzon Ibanez M, Jordana-Ariza N, et al. An update on liquid biopsy analysis for diagnostic and monitoring applications in non-small cell lung cancer. Expert Rev Mol Diagn 2018;18:35-45. [Crossref] [PubMed]
- Pisapia P, Malapelle U, Roma G, et al. Consistency and reproducibility of next-generation sequencing in cytopathology: A second worldwide ring trial study on improved cytological molecular reference specimens. Cancer Cytopathol 2019;127:285-96. [Crossref] [PubMed]
- Malapelle U, Mayo de-Las-Casas C, Rocco D, et al. Development of a gene panel for next-generation sequencing of clinically relevant mutations in cell-free DNA from cancer patients. Br J Cancer 2017;116:802-10. [Crossref] [PubMed]
- Sun S, Thorson JA, Murray SS. Annotation of Variant Data from High-Throughput DNA Sequencing from Tumor Specimens: Filtering Strategies to Identify Driver Mutations. Methods Mol Biol 2019;1908:49-60. [Crossref] [PubMed]
- Gray SW, Hicks-Courant K, Cronin A, et al. Physicians’ attitudes about multiplex tumor genomic testing. J Clin Oncol 2014;32:1317-23. [Crossref] [PubMed]
- Horak P, Frohling S, Glimm H. Integrating next-generation sequencing into clinical oncology: strategies, promises and pitfalls. ESMO Open 2016;1:e000094. [Crossref] [PubMed]
- Kris MG, Johnson BE, Berry LD, et al. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA 2014;311:1998-2006. [Crossref] [PubMed]
- Bailey AM, Mao Y, Zeng J, et al. Implementation of biomarker-driven cancer therapy: existing tools and remaining gaps. Discov Med 2014;17:101-14. [PubMed]
- Weiss GJ. Role and room for biomarkers in non-small cell lung cancer. Precis Cancer Med 2019;2:18. [Crossref]
- Jennings LJ, Arcila ME, Corless C, et al. Guidelines for Validation of Next-Generation Sequencing-Based Oncology Panels: A Joint Consensus Recommendation of the Association for Molecular Pathology and College of American Pathologists. J Mol Diagn 2017;19:341-65. [Crossref] [PubMed]
- Yang H, Wang K. Genomic variant annotation and prioritization with ANNOVAR and wANNOVAR. Nat Protoc 2015;10:1556-66. [Crossref] [PubMed]
- Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010;38:e164. [Crossref] [PubMed]
- Roychowdhury S, Iyer MK, Robinson DR, et al. Personalized oncology through integrative high-throughput sequencing: a pilot study. Sci Transl Med 2011;3:111ra21. [Crossref] [PubMed]
- Parsons HA, Beaver JA, Cimino-Mathews A, et al. Individualized Molecular Analyses Guide Efforts (IMAGE): A Prospective Study of Molecular Profiling of Tissue and Blood in Metastatic Triple-Negative Breast Cancer. Clin Cancer Res 2017;23:379-86. [Crossref] [PubMed]
- Knepper TC, Bell GC, Hicks JK, et al. Key Lessons Learned from Moffitt’s Molecular Tumor Board: The Clinical Genomics Action Committee Experience. Oncologist 2017;22:144-51. [Crossref] [PubMed]
- Castellana S, Fusilli C, Mazza T. A Broad Overview of Computational Methods for Predicting the Pathophysiological Effects of Non-synonymous Variants. Methods Mol Biol 2016;1415:423-40. [Crossref] [PubMed]
- Flanagan SE, Patch AM, Ellard S. Using SIFT and PolyPhen to predict loss-of-function and gain-of-function mutations. Genet Test Mol Biomarkers 2010;14:533-7. [Crossref] [PubMed]
- Molina-Vila MA, Nabau-Moreto N, Tornador C, et al. Activating mutations cluster in the "molecular brake" regions of protein kinases and do not associate with conserved or catalytic residues. Hum Mutat 2014;35:318-28. [Crossref] [PubMed]
- Tafe LJ, Gorlov IP, de Abreu FB, et al. Implementation of a Molecular Tumor Board: The Impact on Treatment Decisions for 35 Patients Evaluated at Dartmouth-Hitchcock Medical Center. Oncologist 2015;20:1011-8. [Crossref] [PubMed]
- Meric-Bernstam F, Farhangfar C, Mendelsohn J, et al. Building a personalized medicine infrastructure at a major cancer center. J Clin Oncol 2013;31:1849-57. [Crossref] [PubMed]
Cite this article as: Molina-Vila MA, Mayo-de -las-Casas C, Garzón-Ibáñez M, Jordana-Ariza N, García-Peláez B, Balada-Bel A, Rosell R. Annotating the next generation sequencing report. Precis Cancer Med 2020;3:6.