
Utilizing genomic profiling in a patient with a thoracic malignancy of uncertain primary: case report
Introduction
Despite advances in diagnostic methods, patients with a diagnosis of cancer of unknown primary (CUP) remain not infrequent and pose significant diagnostic and treatment dilemmas in day to day oncologic clinical practice. CUP is estimated to be 2% of diagnosed cancers (1), although some authors have reported even higher incidences that range from 3% to 10% (2). About two-thirds of cases of CUP are well differentiated or moderately differentiated adenocarcinomas; the remaining third consist of a mix of poorly differentiated carcinomas, squamous cell carcinomas or other poorly differentiated neoplasms (2,3). Main sites of presentation for CUP are liver (40–50%), lymph node (35%), lung (31%), bone metastases (28%), and brain involvement (4,5).
The treatment of choice for patients with unknown tissue of origin has historically been empiric chemotherapy; two drug combinations usually used in first line including a platinum (carboplatin or cisplatin) plus a taxane (paclitaxel or docetaxel) or etoposide, or combinations with one of the previous drugs with gemcitabine or irinotecan (6). Median survival with this type of regimens is estimated to be 9 months, with a 2-year survival rate of 19% (7).
In the era of personalized medicine, better definition of such malignancies is not just feasible but can also be crucial for optimal treatment decisions in order to improve traditional poor outcomes. So far with the improvement of imaging techniques, immunohistochemistry (IHC), and now the use of molecular profiling, management of CUP has improved, but still lags behind advances seen in other areas of oncology.
Case presentation
We present the case of a 62-year-old woman with a past history of hypertension, hyperlipidemia, alcohol dependence, traumatic brain injury after a motor vehicle accident resulting in vocal cord injury and history of a remote stage I left-sided colon adenocarcinoma s/p hemicolectomy in 2008. Patient endorsed ongoing alcohol consumption with 1–3 beers daily and formerly smoked ½ packs per day for 20 years, quit 28 years prior. She denied any family history of cancer.
Patient remained with no evidence of disease on subsequent oncologic follow-up, including a colonoscopy done in 4/2018 which reported no abnormalities. However, in 6/2018 she noted increased hoarseness, 30-pound unintentional weight loss and dysphagia to solids. Work-up showed bulky supraclavicular lymphadenopathy and a massive mediastinal/left lung mass with associated pleural effusion. CT-guided biopsy of the left upper lobe lung lesion was obtained, showing well differentiated adenocarcinoma with extensive necrosis and focal calcifications.
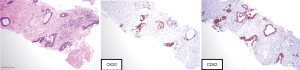
Given location of the mass, patient was initially seen in the thoracic medical oncology clinic with a presumed diagnosis of advanced CUP-possible lung origin. After IHC evaluation (CK7−, CK20+, CDX2+, Figure 1) was more suggestive of a malignancy of lower gastrointestinal origin and genomic profiling by FoundationOne showed two typical Adenomatous Polyposis Coli (APC) mutations frequently encountered in colorectal carcinomas by FoundationOne, her case was reviewed at Molecular Tumor Board and was considered to be very likely a late metastatic recurrence of her colon cancer.
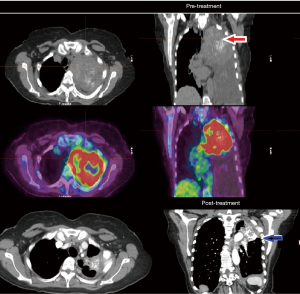
The patient was started on FOLFOX (5-fluorouracil and oxaliplatin) chemotherapy with panitumumab in light of wild-type expanded RAS panel status and in August 2018 with significant clinical improvement (weight gain and resolution of dysphagia) and down trending of tumor markers which was already observed after her second cycle. Patient has continued to have marked clinical, biochemical and objective response which continues now 12 months into therapy (see Figure 2).
Discussion
Isolated recurrence of colon cancer in the thoracic cavity were initially thought to be rare occurrences, documented as case reports (8). This case masquerading as a presumed CUP initially highlights the importance of combining IHC and genomic profiling in cases of poorly differentiated carcinoma and in particular unusual clinical presentations. The patient’s history of colon cancer is highly relevant; there is no recommendation for surveillance imaging for stage I colorectal cancer given the excellent prognosis, as 95% of patients are cured with surgery alone (9,10).
The cascade hypothesis, based on autopsy studies, proposes that colon metastasis occurs in a stepwise fashion; first involving the liver, followed by lung metastases through the liver, and finally arterial metastases (11). The cascade hypothesis makes anatomic sense, but given the cases of isolated metastasis in various anatomic locations, it was challenged by Sadahiro et al., in which they found that frequency of metastasis after curative resection without liver involvement were 39%, and 19% were isolated lung metastasis (12). This favors the hypothesis of hematogenous spread, suggesting that colorectal cancer becomes a systemic disease at an earlier stage, which would fit our case presentation.
It has been documented that approximately 15–20% of patients with CUP benefit from IHC or molecular tests. Identification of a treatment tailored to a specific site of origin is estimated to improve treatment outcomes and disease control might be achieved 30–60% of the time (13).
Several studies have shown success of a variety of types of molecular/expression profiling in CUP. Moran et al. conducted a retrospective analysis by using DNA methylation profiling (panel size of 850 genes—EPICUP) of 216 tissue samples of CUP, predicting the primary site of origin in 188 (87%) of them. Clinical outcomes were available from 92 of the 188 cases profiled by EPICUP. Thirty-one patients received site-specific treatments that matched EPICUP’s predictions, (e.g., 5-fluorouracil for colon carcinoma) and the median overall survival (OS) for this cohort was a favorable 13.6 months. Meanwhile, 61 patients who received empiric chemotherapy not matching the predicted tissue of origin had a median OS of 6 months (14). In another retrospective analysis of tissue samples utilizing a site of origin assay based upon expression profiling by RT-PCR (panel size of 92 genes) by Greco and Hainsworth, 24 of 539 patients initially diagnosed with CUP were re-defined by the assay as renal cell carcinoma (RCC) (4.4%). None of these patients had suspected renal lesions on CT scans. When relatively specific renal IHC stains were performed, 9 of 11 tumors were indeed compatible with RCC. Twenty of 24 patients received RCC specific treatment, achieving a median survival of 16 months (15).
Molecular profiling has also been utilized in prospective studies. Tothill et al. used mRNA microarray next generation sequencing technology (with a 29,285 gene panel) and predicted the primary site in 38 of 49 CUP cases (78%) (5). Hainsworth et al. in another prospective analysis utilizing RT-PCR for mRNA (92 gene panel) predicted the tissue of origin in 247 (98%) of 252 cases. One hundred and ninety-four patients that had tissue of origin prediction received directed therapy, median survival time was 12.5 months. This difference was even more pronounced in patients with treatment responsive tumor types and strong tissue of origin assay prediction with a median survival time of 15.4 months (16). All these studies showed OS outcomes far superior compared to historical results of CUP treated with empiric chemotherapy.
At times there are limitations to obtain a tissue biopsy or the obtained specimen is insufficient for molecular studies (frequently in cases of fine needle aspirations), in which cases recently circulating tumor DNA (ctDNA) technology has led to significant advances and might find further utility in clinical practice. A study evaluated 442 patients with targeted sequencing of 54–70 genes, of which 80% of patients were found to have at least one molecular alteration defined from ctDNA, which is comparable to other tissue-specific studies (17). Nearly all genomic alterations identified were potentially targetable. The benefit of ctDNA is that it can be monitored easily while a patient is receiving therapy and might be helpful to assess treatment response/emerging resistance as well. Furthermore, as more cancer-agnostic therapies become available, such as checkpoint inhibitors for microsatellite instability (MSI)-high tumors and NTRK inhibitors for cancers harboring NTRK fusions, this will become even more relevant. Based on these studies emphasizing role of molecular profiling in work-up of CUP, we modified the current algorithm by European Society for Medical Oncology (ESMO) for management of CUP (Figure 3).

In our case, the patient’s tumor stained negative for CK7, Napsin A, estrogen receptor, Progesterone receptor, PAX8, showed focal immunoreactivity for TTF-1 and was positive for CK20, CDX2, making the case against lung adenocarcinoma, and favoring colorectal adenocarcinoma (18). However, the pattern of a negative CK7 and positive CK 20 is not unique to colorectal adenocarcinoma, as it is also seen in Merkel cell carcinoma, gastric adenocarcinoma (19) and intestinal type of lung adenocarcinoma (20). Treatment for these cancer types are vastly different, which would greatly impact treatment decisions and outcomes. PD-L1 IHC testing showed a tumor proportion score of 0%.
Genomic profiling with FoundationOne was obtained, which has a coding region of over 330 cancer related genes and has a typical median depth of coverage of greater than 250× (21). Results were positive for Tp53 R306, APC Q1338 and APC R876 nonsense mutations. KRAS and NRAS were wildtype. Other notable molecular findings were tumor mutation burden of 4 muts/Mb (low) and microsatellite stable status.
Mutations were reviewed in the Catalogue of Somatic Mutations in Cancer (COSMIC) database (22).
The COSMIC database collects and displays information on somatic mutations in cancer and is the world’s largest database of its kind. It combines two main types of information: high precision manually curated data by experts and genome-wide screen data. Mutation impact used in COSMIC database are derived from the FATHMM-MKL algorithm (23). This is an algorithm that predicts functional, molecular and phenotypic consequences of protein missense variants. Individual mutations from FATHMM-MKL are scored as a P value, ranging from 0 to 1. Deleterious scores are defined above 0.5. COSMIC, however, defines scores over 0.7 as pathogenic to mark their significance.
The gene coding TP53 is the single most common target for genetic insults leading to cancer. DNA damage stabilizes TP53 and allows for TP53 accumulation, which induces tp21 (CDKN1A, CIP1, WAF1) to cause cell cycle arrest (24). When this process is altered, expansion of already existing mutations and additional mutations occur resulting in a malignant tumor. The noted TP53 R306 mutation is a nonsense substitution at position 916 of C->T. It has a pathogenic score of 0.95 by FATHMM (23). This mutation occurs most commonly in tumors of the large intestine (72 samples), breast (36 samples), esophagus (30 samples), stomach (26 samples) and upper aerodigestive tract (23 samples) (22).
The APC gene codes for the APC protein. The function of the APC protein is to bind and regulate degradation of B catenin levels in cytoplasm. When there is an absence of this protein, B catenin levels increase, translocating to nucleus and upregulating cell proliferation (24).
The tumor specimen in our patient’s case harbored two APC mutations. The APC Q1338 mutation is a nonsense substitution in position 4012 of C->T. It has a pathogenic score of 0.90 by FATHMM (23).
According to the COSMIC database, this mutation is found mainly in cancers of the large intestine (71 samples). It is rare in other cancers identified in only one sample of each of the following sites: thyroid, biliary tract, prostate and stomach. The APC p.R876 mutation is also a nonsense substitution mutation in position 2,626 of C->T, tissue distribution also mainly in large intestine (207 samples), with much fewer cases represented in small intestine [4], kidney [3], endometrium [2] and breast [1]. Of note is that neither of these mutations have been reported in thoracic malignancies and biallelic mutations of APC are very proto-typical of colorectal malignancies.
Considering the molecular profiling results of this patient’s tumor, IHC staining and her previous history it was deemed that her current presentation is consistent with metastatic colon adenocarcinoma. It is highly possible that this is a late recurrence of her previous colon adenocarcinoma. The robust response to the treatment regimen directed to RAS-WT colon cancer further corroborates this. While re-testing of the original primary tumor specimen could further validate this, given remote surgery in an outside institution, tissue was not retrievable for such purpose.
Comprehensive genomic profiling proved to be useful in this case as it helped refine diagnosis and guided effective therapy that has been successful to this date. In current guidelines, use of genomic profiling for CUP is viewed as a grade 3 recommendation, arguing that so far it has diagnostic benefit, but not necessarily clinical impact. Our case and emerging data from literature argues that for many patients clinical impact/treatment benefit can also be obtained suggesting potential optimized work flows in the diagnostic work up of CUPs (Figure 3) Further studies will be helpful to guide the implementation of molecular profiling in the management of CUP.
We presented this case in accordance with the CARE-Guideline (25).
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editor Balazs Halmos for the series “Precision Oncology Tumor Board” published in Precision Cancer Medicine. The article has undergone external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/pcm.2019.11.05). The Series “Precision Oncology Tumor Board” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and national research committees and with the Helsinki Declaration (as revised in 2013). Written informed consent was obtained from the patient for publication of this manuscript and any accompanying images.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Hainsworth JD, Greco FA. Cancer of Unknown Primary Site: New Treatment Paradigms in the Era of Precision Medicine. Am Soc Clin Oncol Educ Book 2018;38:20-5. [Crossref] [PubMed]
- Moran S, Martinez-Cardus A, Boussios S, et al. Precision medicine based on epigenomics: the paradigm of carcinoma of unknown primary. Nat Rev Clin Oncol 2017;14:682-94. [Crossref] [PubMed]
- Pentheroudakis G, Golfinopoulos V, Pavlidis N. Switching benchmarks in cancer of unknown primary: from autopsy to microarray. Eur J Cancer 2007;43:2026-36. [Crossref] [PubMed]
- Pavlidis N, Pentheroudakis G. Cancer of unknown primary site. Lancet 2012;379:1428-35. [Crossref] [PubMed]
- Tothill RW, Shi F, Paiman L, et al. Development and validation of a gene expression tumour classifier for cancer of unknown primary. Pathology 2015;47:7-12. [Crossref] [PubMed]
- NCCN Guidelines Version 1.2020 Occult Primary [Internet]. NCCN. 2019 [cited NCCN Guidelines Occult Primary]. Available online: https://www.nccn.org/professionals/physician_gls/pdf/occult.pdf
- Greco FA, Pavlidis N. Treatment for patients with unknown primary carcinoma and unfavorable prognostic factors. Semin Oncol 2009;36:65-74. [Crossref] [PubMed]
- Daniels AM, Vogelaar JFJ. Late onset pulmonary metastasis more than 10 years after primary sigmoid carcinoma. World J Gastrointest Pathophysiol 2017;8:96-9. [Crossref] [PubMed]
- Seo SI, Lim SB, Yoon YS, et al. Comparison of recurrence patterns between ≤5 years and >5 years after curative operations in colorectal cancer patients. J Surg Oncol 2013;108:9-13. [Crossref] [PubMed]
- NCCN Guidelines Version 3.2019 Colon Cancer [Internet]. 2019. Available online: https://www.nccn.org/professionals/physician_gls/pdf/colon.pdf
- Weiss L, Grundmann E, Torhorst J, et al. Haematogenous metastatic patterns in colonic carcinoma: an analysis of 1541 necropsies. J Pathol 1986;150:195-203. [Crossref] [PubMed]
- Sadahiro S, Suzuki T, Tanaka A, et al. Hematogenous metastatic patterns of curatively resected colon cancer were different from those of stage IV and autopsy cases. Jpn J Clin Oncol 2013;43:444-7. [Crossref] [PubMed]
- Collado Martin R, Garcia Palomo A, de la Cruz Merino L, et al. Clinical guideline SEOM: cancer of unknown primary site. Clin Transl Oncol 2014;16:1091-7. [Crossref] [PubMed]
- Moran S, Martinez-Cardus A, Sayols S, et al. Epigenetic profiling to classify cancer of unknown primary: a multicentre, retrospective analysis. Lancet Oncol 2016;17:1386-95. [Crossref] [PubMed]
- Greco FA, Hainsworth JD. Renal Cell Carcinoma Presenting as Carcinoma of Unknown Primary Site: Recognition of a Treatable Patient Subset. Clin Genitourin Cancer 2018;16:e893-8. [Crossref] [PubMed]
- Hainsworth JD, Rubin MS, Spigel DR, et al. Molecular gene expression profiling to predict the tissue of origin and direct site-specific therapy in patients with carcinoma of unknown primary site: a prospective trial of the Sarah Cannon research institute. J Clin Oncol 2013;31:217-23. [Crossref] [PubMed]
- Kato S, Krishnamurthy N, Banks KC, et al. Utility of Genomic Analysis In Circulating Tumor DNA from Patients with Carcinoma of Unknown Primary. Cancer Res 2017;77:4238-46. [Crossref] [PubMed]
- Kummar S, Fogarasi M, Canova A, et al. Cytokeratin 7 and 20 staining for the diagnosis of lung and colorectal adenocarcinoma. Br J Cancer 2002;86:1884-7. [Crossref] [PubMed]
- Selves J, Long-Mira E, Mathieu MC, et al. Immunohistochemistry for Diagnosis of Metastatic Carcinomas of Unknown Primary Site. Cancers (Basel) 2018; [Crossref] [PubMed]
- Stojsic J, Kontic M, Subotic D, et al. Intestinal type of lung adenocarcinoma in younger adults. Case Rep Pulmonol. 2014;2014:282196. [Crossref] [PubMed]
- Frampton GM, Fichtenholtz A, Otto GA, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol 2013;31:1023-31. [Crossref] [PubMed]
- Tate JG, Bamford S, Jubb HC, et al. COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res 2019;47:D941-7. [Crossref] [PubMed]
- Rogers MF, Shihab HA, Mort M, et al. FATHMM-XF: accurate prediction of pathogenic point mutations via extended features. Bioinformatics 2018;34:511-3. [Crossref] [PubMed]
- Kumar V, Abbas AK, Aster JC. Robbins Basic Pathology. 10th edition. Elsevier, 2018.
- Riley DS, Barber MS, Kienle GS, et al. CARE 2013 Explanations and Elaborations: Reporting Guidelines for Case Reports. J Clin Epidemiol 2017;89:218-35. [Crossref] [PubMed]
Cite this article as: Galeas JN, Shah N, Fidvi SA, Chuy J. Utilizing genomic profiling in a patient with a thoracic malignancy of uncertain primary: case report. Precis Cancer Med 2020;3:9.

