替代非同源末端连接修复:癌症基因组不稳定性的主要调节器
引言
基因组不稳定性-癌症特征之一
基因组不稳定性是指在基因组内积累遗传缺陷的趋势的生物事件。由于其促进有害突变,基因组的不稳定性逐渐成为驱动肿瘤发生和进展的特征[1,2]。基因组不稳定性包括以下几类:核苷酸不稳定性(NIN),其特征为碱基缺失、插入或替换[3];微卫星不稳定性(MSI),以微卫星区域寡核苷酸重复序列的扩张或收缩为特征[4,5];染色体不稳定性,这也是最具代表性的基因组不稳定性类型,以数值或结构染色体变异为特征[6,7]。
关于基因组不稳定性的评估,如某些类型的易位或缺失,与肿瘤患者的预后相关[8,9]。近期已被证实:基因组不稳定性可以作为一个重要的预测因子,用于评估同源重组(HR)缺陷或MSI情况下应用PD1抑制剂[11],通过合成致死方法选择性地靶向肿瘤细胞[10]。因此,努力探索基因组不稳定性的潜在机制将成为肿瘤精准治疗中一个重要的挑战。
DNA修复途径:概述
尽管基因组不稳定性的分子机制在很大程度上尚不为人所知,但越来越多的证据提示DNA修复机制的紊乱可能发挥关键作用,因为它可以容忍DNA损伤超载,从而选择具有增殖和存活优势的细胞[12]。
人类细胞持续接收大量(大约7万/天)DNA损伤[13]。DNA损伤既可能来自氧化损伤、复制叉崩溃和端粒受损等内源因素,又可能来自电离辐射(IR)、紫外线或化学暴露等外源性因素。大多数损伤是单链DNA(ssDNA)断裂,这些损伤可随机转化为更危险的DNA双链断裂(DSBs)。然而,在免疫球蛋白多样性和功能生成过程中,V(D)J重组和类开关重组(CSR)等生理事件也会产生DSBs。
为了保持基因组的稳定性,几种DNA修复途径共同协作以避免未修复的双链断裂造成的影响。事实上,如果DSBs过广,正常细胞会激活细胞凋亡程序来阻止受损后代细胞的增殖。然而在肿瘤细胞中,DSB经常被错误地修复,导致基因组不稳定,从而可能导致癌变和进展。
DSB修复的两个主要途径是经典的非同源端连接(C-NHEJ)和HR。
NHEJ可发生在细胞周期的所有阶段,因此是DSBs修复的主要途径[15]。C-NHEJ修复与断端切除无关,是一个非常快速的过程。DSB识别由Ku70-Ku80异源二聚体介导,它与DNA 末端结合,然后招募DNA依赖的蛋白激酶催化亚基(DNA-pkcs)来磷酸化Artemis,进而处理单链悬臂。最终DNA连接酶4(LIG4)与支架蛋白XRCC4催化DNA末端的连接。尽管C-NHEJ在没有同源DNA模板的情况下修复DSBs,DSB识别由Ku70-Ku80异源二聚体操作,它与DNA 末端结合,然后招募DNA依赖的蛋白激酶催化亚基(DNA-pkcs)来磷酸化Artemis,进而处理单链悬臂。最后,DNA连接酶4(LIG4)和支架蛋白XRCC4催化DNA末端的连接。虽然C-NHEJ在没有同源DNA模板的情况下修复dsb,但它的高效率只与连接处有限的序列改变有关,这使得该途径确保了基因组稳定性[16]。
和C-NHEJ不同,HR在细胞周期的S期和G2期阶段发挥作用,因为它需要同源DNA模板来恢复DSB周围的序列[17]。第一步是DNA末端切除,该5'端的核苷酸被消化并产生长3'端单链DNA(ssDNA)。这一过程由Mre11,Rad50,Nbs1(MRN化合物)和CtIP、BRCA1等辅助蛋白完成,这些蛋白可以感知DSBs,启动HR机制,向DSB招募。3'-ssDNA尾部被复制蛋白A(RPA)复合物和Rad51核蛋白丝包裹并稳定,随后链侵入相邻完整的姐妹染色单体和形成D-环。最后,聚合酶催化DNA合成,直到Holliday连接被分解和DSB修复[18]。
除了C-NHEJ和HR,最近的实验证据表明还存在第三种DSB修复途径,该途径被命名为Alt-NHEJ[19-21]。
在这篇评论中,我们重点关注Alt-NHEJ修复在促进基因组不稳定性中的作用,在这篇综述中,我们重点关注Alt-NHEJ修复在促进基因组不稳定性中的作用,强调其作为个性化治疗创新治疗靶点的潜在价值。
肿瘤中的Alt-NHEJ负调节
Alt-NHEJ机制
Alt-NHEJ修复包括三个子途径,DSBs DNA末端的DNA序列的互补程度决定了具体的修复途径。微同源介导的末端连接(MMEJ)需要2-20个同源序列的核苷酸[21];单链退火(SSA)需要同源序列的核苷酸大于25个;第三个子途径是特征不明显的末端连接(EJ),它几乎不需要修复位上有同源序列。
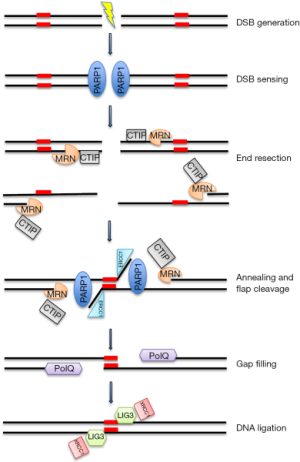
我们将以MMEJ为例来描述Alt-NHEJ,因为它是更好描述的子途径(图1)。
尽管大多数的Alt-NHEJ修复发生在细胞周期的S和G2阶段,它也在G1阶段发生[22]。第一步需要通过MRN/CtIP复合物进行DNA末端切除[23],该复合物作为DNA不连续的感应器被聚ADP核糖聚合酶-1(PARP-1)招募到DSB上[24]。特别的是,CtIP激活增强了MRN内切酶/外切酶的活性[25],导致单链区域内微同源序列的显示。下一步是通过PARP-1、MRN和PolQ共同作用的短微同源性对DNA末端进行桥接和对齐[26,27]。然后,非同源3'尾部被ERCC1\XPF核酸酶消化,在DNA链中产生缺口,该缺口由PolQ为介质的DNA合成物填补[27]。最后,DNA DSBs最终被DNA连接酶3(LIG3)/XRCC1复合物修复。特别是支架蛋白XRCC1通过与PARP-1和MRN复合物的物理作用驱动靠近DNA断裂处附近的LIG3,最终连接DSB[28,29]。虽然LIG3被认为在DNA连接的最后一步更有效,但在没有LIG3的情况下,DNA连接酶1(LIG1)也可发挥这一功能[30]。
与其他DSB修复途径的关系
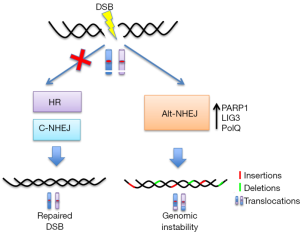
一些研究认为Alt-NHEJ是在C-NHEJ或HR缺失时的后备途径[31](图2)。尽管不同的机制已提示,当C-NHEJ或HR机制没能修复DNA末端时,Alt-NHEJ可能会接替DSB修复:
- DSB感知:DNA断裂感应器PARP-1和Ku可以相互竞争修复DSB[32],而Ku对末端更高的亲和力可以解释C-NHEJ的优势。Ku的下调可导致Alt-NHEJ活性增加。
- DNA末端加工:CDKs磷酸化CtIP[33]或SIRT6去乙酰化CtIP可增强Alt-NHEJ[34],而53BP1阻止CtIP通过C-NHEJ介导DNA末端直接修复[22]。
- 空隙填充:PolQ具有一个rad51结合域,可抑制HR从而促进AL T-NHEJ[35];此外,polθ-解旋酶活性有助于从切除的dsb中去除RPA,使其退火并随后由Alt-NHEJ连接[36]。
- DNA连接:lig4缺失细胞的DNA末端连接修复由lig3驱动的AltNHEJ补偿[30]。
除了作为后备途径外,我们推测Alt-NHEJ还有其他的生理作用。最近的一项研究表明,Alt-NHEJ在某些末端受损的DSBs上被激活,比如由IR诱导的那些[37]。此外,因为LIG3是线粒体的主要DNA连接酶,Alt-NHEJ修复在线粒体DNA(mtDNA)的DSB修复中起着关键作用。
Alt-NHEJ与基因组不稳定性
Alt-NHEJ被认为是一种容易出错的DNA修复途径,原因有以下几点:(1)由于缺少模板链,在DSB附近无法正确恢复原始DNA序列[31];(2)不精确的末端连接机制,其特征是混乱的调控因素和延迟修复动力学改变[19];(3)由于末端转移酶活性和ssDNA的迭代合成,通过PoIQ进行低精确度的缺口填充,进而在修复点产生显著的序列畸变(插入)[39,40];(4)核酸内切/外切酶机制产生的大量基因缺失以揭示微同源区域[41];(5)由负责产生易位的LIG3的N端锌指结构域(ZnF),连接不相关的DNA分子[43]。这些基因组畸变通常是由V(D)J和CSR重组过程的错误引起,其中程序性DSB主要通过C-NHEJ途径重新连接,C-NHEJ途径是基因组不稳定性的抑制因子[44-46]。C-NHEJ缺陷的小鼠(因为缺乏Ku80、XRCC4、连接酶IV、DNA-PKcs或Artemis)也有P53缺陷,这些小鼠通常会发展为包含癌性染色体易位的前B细胞淋巴瘤,包含IgH和c-Myc位点[47],而所有这些都由Alt-NHEJ催化。此外,任何C-NHEJ蛋白有缺陷的SCID小鼠[48,49]均易发生肿瘤,涉及Alt-NHEJ诱发产生Ig位点。相反地,缺少Alt-NHEJ核心成分PARP-1和LIG3的小鼠细胞,或经PARP抑制剂处理过的小鼠细胞染色体易位的总体频率降低,因而基因组不稳定性的水平较低[43,50]。
Alt-NHEJ抑制策略
因为Alt-NHEJ主要作为HR或NHEJ这两个主要的DBS修复途径的后备途径,因此其是在HR或NHEJ缺陷性肿瘤中的一个很有前景的治疗靶点。事实上,抑制Alt-NHEJ只会杀死依赖于Alt-NHEJ修复路径来修复DNA受损的肿瘤细胞而保留正常细胞。
目前抑制Alt-NHEJ可通过阻断以下途径来实现。(1)通过PARP-1/2抑制剂检测DNA传感的早期过程。(2)通过DNA连接酶LIG1/LIG3抑制剂来实现DNA末端连接的最后一步。
①PARP1是PARP家族中特征最明显的成员。它通过其DNA结合区域感知DNA损伤,随后合成聚(ADP-核糖)(PAR),并添加到自身和其他受体蛋白中。由此产生的PARylation招募了包括LIG3和XRCC1在内的其他DNA修复蛋白[51,52]。
PARP抑制剂通过与NAD+竞争PARP的催化亚基来发挥其活性[53]。PARP抑制剂在缺乏HR的肿瘤细胞内诱导合成致死细胞死亡的证据[54,55]已得到了不同的作用机制的支持[56]。由于PARP1参与BER介导的修复,现有大部分模型已表明,正常细胞活动中产生的DNA SSBs在PARP抑制剂处理后仍然存在,并可在HR缺陷的细胞内转化为DSBs,诱导细胞死亡。然而,PARP抑制剂治疗期间SSB积累的缺失是对PARP抑制剂治疗HR缺陷肿瘤细胞中合成致死现象的另一种解释[57]。在这方面,一个有趣的模型表明,HR缺陷细胞高度依赖于Alt-NHEJ后备通路,该通路被PARP抑制剂阻断,导致DNA损伤超载和凋亡细胞死亡[35]。
与这一临床前证据一致,美国食品和药品监督管理局(FDA)批准了三种PARP抑制剂(olaparib、rucaparib和niraparib)用于治疗卵巢癌(特别是复发的铂敏感性高级别浆液性疾病,包括gBRCAm),以及olaparib用于治疗gBRCAm乳腺癌。
然而,目前尚存在一些挑战,包括识别预测性生物标志物,如HR修复缺陷信号,以精确识别可能从PARPi单独或与化疗、靶向药物、放疗或免疫治疗联合中获益的患者亚群。
②DNA连接酶是ATP依赖酶,可催化DNA链的末端连接。催化核心在缺失DNA时采用扩展的构象,在DNA损伤时在DNA缺口周围形成环状结构。LIG3与LIG1和LIG4有不同的区别。首先,LIG3基因的替代翻译起始会产生两种具有不同细胞功能和定位的亚型[58]:a.在切除修复过程和Alt-NHEJ修复中起作用的核LIG3[59];b.参与线粒体DNA代谢的线粒体LIG3[38,60]。其次,LIG3具有特定的N端ZnF,其在感知DNA断裂和DNA分子间连接中起着关键的作用[42]。
Chen等人发现了3种通过阻断DBD来抑制DNA连接酶的小分子。其中,L82抑制LIG1,L67抑制LIG1和LIG3,L189抑制DNA连接酶I、III和IV。重要的是,DNA连接酶抑制剂发挥细胞毒性或细胞抑制(L82)作用,并使癌细胞对DNA损伤剂敏感[61]。此外,最近Sallmyr等研究表明,L67在肿瘤细胞中优先靶向线粒体LIG3功能,进而增加核DNA损伤和细胞死亡,而在非恶性细胞中没有任何显著影响[62]。
Alt-NHEJ作为肿瘤的治疗靶点:临床前研究
在接下来的章节里,我们将描述Alt-NHEJ在某些肿瘤的发病、进展和耐药性中的作用。
卵巢癌
大约一半的上皮性卵巢癌(EOCs)存在HR修复缺陷,这驱动了基因组不稳定性和对PARP定向DNA修复的依赖。Ceccaldi等报道在EOCs患者中HR活性与PoIQ表达呈负相关[35]。此外,HR缺陷的肿瘤细胞具有更高的稳定水平的PoIQ,其是参与Alt-NHEJ的DNA聚合酶。此外,作者还发现PoIQ本身可以通过阻断RAD51介导的重组来抑制HR。重要的是,在HR缺陷的EOCs中敲低PoIQ会降低细胞存活率,而在小鼠中FANCD2/BRCA2和PoIQ的遗传失活会导致胚胎致死。总的来说,这些结果证实了在EOCs中HR途径与Alt-NHEJ修复之间的合成致命关系,表明PoIQ是HR缺陷肿瘤的一个新的潜在治疗靶点。
乳腺癌
Alt-NHEJ与乳腺癌发病和获得性耐药有关。Tobin等人证明雌激素受体和孕激素受体阳性(ER/PR+)MCF7乳腺癌细胞与非致瘤性乳腺上皮MCF10A细胞相比,Alt-NHEJ通路活性增加。此外,作者还发现MCF7细胞中抗他莫昔芬和芳香化酶衍生物的核心成分可上调LIG3和PARP1,从而增加对PARP和LIG3抑制剂组合的敏感性[63]。
神经母细胞瘤
Newman等人发现Alt-NHEJ对神经母细胞瘤的基因组不稳定性和细胞存活至关重要[64]。事实上,他们证明了一个容易出错的DNA修复通路在神经母细胞瘤细胞中的过度激活。与观察到的Alt-NHEJ和PARP1所需的蛋白上调相比,该通路表现为低水平的C-NHEJ、LIG4和Artemis(DCLRE1C)介质,特别是在MYCN过表达细胞株中。通过L67和BYK204165(BYK)来分别抑制LIG3和PARP1,可导致DSB积累和细胞死亡。通过数据集分析显示:高水平编码Alt-NHEJ蛋白与总体低生存率相关。此外,作者首次表明,Alt-NHEJ通过介导人神经嵴干细胞分化中的MYCN致癌活性,参与神经细母胞瘤的起始[65]。
白血病
有实验证据表明,TK活化白血病的特征是基因组深度不稳定性。
Fan等报道在表达FLT3/ITD的急性骨髓性白血病细胞株和FLT3/ITD敲入小鼠的骨髓单核细胞中,Alt-NHEJ修复过度激活,导致DNA序列高频率畸变。在FLT3/ITD表达细胞中,C-NHEJ Ku核心成分蛋白水平降低,LIG3表达上调。重要的是,作者发现,在FLT3/ITD表达细胞中,FLT3信号可增强Alt-NHEJ修复活性,并且使用FLT3抑制剂(CEP-701)能够降低LIG3介导的基因组不稳定性[66]。此外,Hähnel等人证实:致癌的KRAS导致T细胞急性淋巴母细胞性白血病(T-ALL)中Alt-NHEJ通路的成分上调。重要的是,靶向Alt-NHEJ通路可选择性地使KRAS突变的白血病细胞对细胞毒性药物,如阿糖胞苷、柔红霉素或VP-16敏感[67]。有意思的是,Muvarak等人发现c-MYC通过增加易于出错的Alt-NHEJ修复的关键成分LIG3和PARP1的表达,促进了TK激活白血病中观察到的基因组不稳定性[68]。
Sallmyr等人也发现慢性髓系白血病(CML) BCR-ABL阳性细胞的Alt-NHEJ修复活性增加。事实上,C-NHEJ通路的关键蛋白Artemis和LIG4表达下调,而LIG3表达上调。值得注意的是,敲低LIG3可导致DNA损伤增加,从而潜在地影响了CML细胞的存活[69]。
多发性骨髓瘤(MM)
MM是一种血液恶性肿瘤,其特征是浆细胞异常增殖,并伴有多种染色体畸变。我们的研究表明,LIG3介导的DNA修复上调在MM细胞的基因组不稳定性和存活中发挥关键作用[70]。特别是,在MM患者中,LIG3 mRNA的高表达水平与预后差相关。在体内和体外实验均证实,LIG3的持续敲低会抑制MM细胞的活力,这表明恶性浆细胞依赖于LIG3驱动的修复。重要的是,我们发现LIG3的表达受miR-22的调控。事实上,MM细胞中miR-22类似物可下调LIG3蛋白,减少Alt-NHEJ修复,增加DNA损伤和细胞死亡。总的来说,这些结果表明:骨髓瘤细胞对LIG3驱动的Alt-NHEJ修复成瘾,调节MM的发生发展和耐药性。
此外,针对各个疾病阶段患者的靶向测序研究结果显示,MM样本中同源重组缺陷(HRD)与疾病的发展和耐药获得呈现正相关,并与高危分层标志物相关[71]。由于Alt-NHEJ可以弥补HR缺陷修复,这些数据为评估PARP抑制剂在MM患者中的作用提供了依据,特别是在复发的情况下。事实上,我们的初步结果表明,Alt-NHEJ抑制剂对耐药MM细胞株和原始初级细胞具有高度活性(Caracciolo等人,手稿准备中),这也进一步证实了ALt-NHEJ与MM发病机制的相关性。
结论和展望
虽然DDR的失调代表了一种有害的机制,肿瘤细胞通过这种机制可获得突变体表型,但它同时也为抗肿瘤治疗提供了新思路。事实上,肿瘤细胞通常具有至少有一条不受控的DNA修复通路,而这一缺陷通过激活第二条通路得到补偿,第二条通路的抑制将特异性地杀死肿瘤细胞(合成致死性),而对正常细胞几乎没有影响。因此,这种策略为设计新的治疗方法提供了思路,可以选择性地杀死肿瘤细胞,提高疾病反应,并使患者免于不必要的不良反应。然而,为探索肿瘤生物行为的其他潜在缺陷,在不久的将来,DDR缺陷的生物标志物的识别会成为一个挑战。
最近的研究表明:PARP抑制剂在BRCA缺陷卵巢癌和乳腺癌中的疗效,这一现象提示存在HR缺陷或NHEJ缺陷的肿瘤细胞更依赖于Alt-NHEJ DNA修复机制,也为基于Alt-NHEJ设计的治疗策略提供了很好的依据。事实上,在这篇综述中,我们已描述了在DSB修复途径缺陷的几类实体瘤和血液恶性肿瘤中的核心成分,如PARP-1、LIG3和PolQ表达升高,这可能为Alt-NHEJ抑制剂应答提供预测性的生物标志物。
虽然DNA修复基因的改变不能解释所有基因组不稳定的病例,特别是在散发性肿瘤中,但我们报告了Alt-NHEJ在大型基因组重排(特别是易位)中发挥关键作用的证据,这是不同肿瘤发病和发展的基础。
由于NGS技术允许在个体患者中描述个体DNA修复途径的状态[72],在Alt-NHEJ上调的肿瘤中,Alt-NHEJ抑制剂与其他治疗模式(如化疗、免疫治疗或放疗)相结合,可能是一个有前途的治疗策略。
总之,本综述强调易出错的Alt-NHEJ修复途径是HR或NHEJ缺陷肿瘤的阿喀琉斯之踵,合成致死策略有望在精准肿瘤学的临床应用中加以利用。
Acknowledgments
Funding: This work has been mainly supported by the Italian Association for Cancer Research (AIRC) with “Special Program for Molecular Clinical Oncology 5 per mille”, 2010/15 and its Extension Program” No. 9980, 2016/18 (PI: PT); and also by “Innovative Immunotherapeutic Treatments of Human Cancer” Multi Unit Regional No. 16695 (co-financed by AIRC and the CARICAL foundation). We thanks Dr. Ivana Criniti for her study coordination support and editorial assistance.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editor Masood A. Shammas for the series “Genomic Instability, Clonal Evolution and Oncogenesis” published in Precision Cancer Medicine. The article has undergone external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/pcm.2019.02.05). The series “Genomic Instability, Clonal Evolution and Oncogenesis” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability--an evolving hallmark of cancer. Nat Rev Mol Cell Biol 2010;11:220-8. [Crossref] [PubMed]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646-74. [Crossref] [PubMed]
- Dworaczek H, Xiao W. Xeroderma pigmentosum: a glimpse into nucleotide excision repair, genetic instability, and cancer. Crit Rev Oncog 2007;13:159-77. [Crossref] [PubMed]
- De' Angelis GL. Microsatellite instability in colorectal cancer. Acta Biomed 2018;89:97-101. [PubMed]
- Abida W, Cheng ML, Armenia J, et al. Analysis of the Prevalence of Microsatellite Instability in Prostate Cancer and Response to Immune Checkpoint Blockade. JAMA Oncol 2018; [Epub ahead of print]. [Crossref] [PubMed]
- Sheffer M, Bacolod MD, Zuk O, et al. Association of survival and disease progression with chromosomal instability: a genomic exploration of colorectal cancer. Proc Natl Acad Sci U S A 2009;106:7131-6. [Crossref] [PubMed]
- Fenech M. Chromosomal biomarkers of genomic instability relevant to cancer. Drug Discov Today 2002;7:1128-37. [Crossref] [PubMed]
- Sansregret L, Nepveu A. Gene signatures of genomic instability as prognostic tools for breast cancer. Future Oncol 2011;7:591-4. [Crossref] [PubMed]
- Fonseca R, Blood E, Rue M, et al. Clinical and biologic implications of recurrent genomic aberrations in myeloma. Blood 2003;101:4569-75. [Crossref] [PubMed]
- Murata S, Zhang C, Finch N, et al. Predictors and Modulators of Synthetic Lethality: An Update on PARP Inhibitors and Personalized Medicine. Biomed Res Int 2016;2016:2346585. [Crossref] [PubMed]
- Asaoka Y, Ijichi H, Koike K. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med 2015;373:1979. [Crossref] [PubMed]
- Jeggo PA, Pearl LH, Carr AM. DNA repair, genome stability and cancer: a historical perspective. Nat Rev Cancer 2016;16:35-42. [Crossref] [PubMed]
- Lindahl T, Barnes DE. Repair of endogenous DNA damage. Cold Spring Harb Symp Quant Biol 2000;65:127-33. [Crossref] [PubMed]
- Hoeijmakers JH. DNA damage, aging, and cancer. N Engl J Med 2009;361:1475-85. [Crossref] [PubMed]
- Pannunzio NR, Watanabe G, Lieber MR. Nonhomologous DNA end-joining for repair of DNA double-strand breaks. J Biol Chem 2018;293:10512-23. [Crossref] [PubMed]
- Burma S, Chen BP, Chen DJ. Role of non-homologous end joining (NHEJ) in maintaining genomic integrity. DNA Repair (Amst) 2006;5:1042-8. [Crossref] [PubMed]
- San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem 2008;77:229-57. [Crossref] [PubMed]
- Heyer WD, Ehmsen KT, Liu J. Regulation of homologous recombination in eukaryotes. Annu Rev Genet 2010;44:113-39. [Crossref] [PubMed]
- Wang H, Perrault AR, Takeda Y, et al. Biochemical evidence for Ku-independent backup pathways of NHEJ. Nucleic Acids Res 2003;31:5377-88. [Crossref] [PubMed]
- Terzoudi GI, Singh SK, Pantelias GE, et al. Premature chromosome condensation reveals DNA-PK independent pathways of chromosome break repair. Int J Oncol 2008;33:871-9. [PubMed]
- Chang HHY, Pannunzio NR, Adachi N, et al. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat Rev Mol Cell Biol 2017;18:495-506. [Crossref] [PubMed]
- Xiong X, Du Z, Wang Y, et al. 53BP1 promotes microhomology-mediated end-joining in G1-phase cells. Nucleic Acids Res 2015;43:1659-70. [Crossref] [PubMed]
- Xie A, Kwok A, Scully R. Role of mammalian Mre11 in classical and alternative nonhomologous end joining. Nat Struct Mol Biol 2009;16:814-8. [Crossref] [PubMed]
- Yang G, Liu C, Chen SH, et al. Super-resolution imaging identifies PARP1 and the Ku complex acting as DNA double-strand break sensors. Nucleic Acids Res 2018;46:3446-57. [Crossref] [PubMed]
- Anand R, Ranjha L, Cannavo E, et al. Phosphorylated CtIP Functions as a Co-factor of the MRE11-RAD50-NBS1 Endonuclease in DNA End Resection. Mol Cell 2016;64:940-50. [Crossref] [PubMed]
- Haince JF, McDonald D, Rodrigue A, et al. PARP1-dependent kinetics of recruitment of MRE11 and NBS1 proteins to multiple DNA damage sites. J Biol Chem 2008;283:1197-208. [Crossref] [PubMed]
- Kent T, Chandramouly G, McDevitt SM, et al. Mechanism of microhomology-mediated end-joining promoted by human DNA polymerase theta. Nat Struct Mol Biol 2015;22:230-7. [Crossref] [PubMed]
- Okano S, Lan L, Tomkinson AE, et al. Translocation of XRCC1 and DNA ligase IIIalpha from centrosomes to chromosomes in response to DNA damage in mitotic human cells. Nucleic Acids Res 2005;33:422-9. [Crossref] [PubMed]
- Della-Maria J, Zhou Y, Tsai MS, et al. Human Mre11/human Rad50/Nbs1 and DNA ligase IIIalpha/XRCC1 protein complexes act together in an alternative nonhomologous end joining pathway. J Biol Chem 2011;286:33845-53. [Crossref] [PubMed]
- Lu G, Duan J, Shu S, et al. Ligase I and ligase III mediate the DNA double-strand break ligation in alternative end-joining. Proc Natl Acad Sci U S A 2016;113:1256-60. [Crossref] [PubMed]
- Iliakis G, Murmann T, Soni A. Alternative end-joining repair pathways are the ultimate backup for abrogated classical non-homologous end-joining and homologous recombination repair: Implications for the formation of chromosome translocations. Mutat Res Genet Toxicol Environ Mutagen 2015;793:166-75. [Crossref] [PubMed]
- Paddock MN, Bauman AT, Higdon R, et al. Competition between PARP-1 and Ku70 control the decision between high-fidelity and mutagenic DNA repair. DNA Repair (Amst) 2011;10:338-43. [Crossref] [PubMed]
- Barton O, Naumann SC, Diemer-Biehs R, et al. Polo-like kinase 3 regulates CtIP during DNA double-strand break repair in G1. J Cell Biol 2014;206:877-94. [Crossref] [PubMed]
- Mao Z, Hine C, Tian X, et al. SIRT6 promotes DNA repair under stress by activating PARP1. Science 2011;332:1443-6. [Crossref] [PubMed]
- Ceccaldi R, Liu JC, Amunugama R, et al. Homologous-recombination-deficient tumours are dependent on Poltheta-mediated repair. Nature 2015;518:258-62. [Crossref] [PubMed]
- Mateos-Gomez PA, Kent T, Deng SK, et al. The helicase domain of Poltheta counteracts RPA to promote alt-NHEJ. Nat Struct Mol Biol 2017;24:1116-23. [Crossref] [PubMed]
- Dutta A, Eckelmann B, Adhikari S, et al. Microhomology-mediated end joining is activated in irradiated human cells due to phosphorylation-dependent formation of the XRCC1 repair complex. Nucleic Acids Res 2017;45:2585-99. [PubMed]
- Simsek D, Furda A, Gao Y, et al. Crucial role for DNA ligase III in mitochondria but not in Xrcc1-dependent repair. Nature 2011;471:245-8. [Crossref] [PubMed]
- Arana ME, Seki M, Wood RD, et al. Low-fidelity DNA synthesis by human DNA polymerase theta. Nucleic Acids Res 2008;36:3847-56. [Crossref] [PubMed]
- Wood RD, Doublie S. DNA polymerase theta (POLQ), double-strand break repair, and cancer. DNA Repair (Amst) 2016;44:22-32. [Crossref] [PubMed]
- Zhuang J, Jiang G, Willers H, et al. Exonuclease function of human Mre11 promotes deletional nonhomologous end joining. J Biol Chem 2009;284:30565-73. [Crossref] [PubMed]
- Taylor RM, Whitehouse CJ, Caldecott KW. The DNA ligase III zinc finger stimulates binding to DNA secondary structure and promotes end joining. Nucleic Acids Res 2000;28:3558-63. [Crossref] [PubMed]
- Simsek D, Brunet E, Wong SY, et al. DNA ligase III promotes alternative nonhomologous end-joining during chromosomal translocation formation. PLoS Genet 2011;7:e1002080. [Crossref] [PubMed]
- Panchakshari RA, Zhang X, Kumar V, et al. DNA double-strand break response factors influence end-joining features of IgH class switch and general translocation junctions. Proc Natl Acad Sci U S A 2018;115:762-7. [Crossref] [PubMed]
- Kotnis A, Du L, Liu C, et al. Non-homologous end joining in class switch recombination: the beginning of the end. Philos Trans R Soc Lond B Biol Sci 2009;364:653-65. [Crossref] [PubMed]
- Yan CT, Boboila C, Souza EK, et al. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature 2007;449:478-82. [Crossref] [PubMed]
- Zhu C, Mills KD, Ferguson DO, et al. Unrepaired DNA breaks in p53-deficient cells lead to oncogenic gene amplification subsequent to translocations. Cell 2002;109:811-21. [Crossref] [PubMed]
- Zhu C, Bogue MA, Lim DS, et al. Ku86-deficient mice exhibit severe combined immunodeficiency and defective processing of V(D)J recombination intermediates. Cell 1996;86:379-89. [Crossref] [PubMed]
- Taccioli GE, Amatucci AG, Beamish HJ, et al. Targeted disruption of the catalytic subunit of the DNA-PK gene in mice confers severe combined immunodeficiency and radiosensitivity. Immunity 1998;9:355-66. [Crossref] [PubMed]
- Wray J, Williamson EA, Singh SB, et al. PARP1 is required for chromosomal translocations. Blood 2013;121:4359-65. [Crossref] [PubMed]
- Krishnakumar R, Kraus WL. The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets. Mol Cell 2010;39:8-24. [Crossref] [PubMed]
- Morales J, Li L, Fattah FJ, et al. Review of poly (ADP-ribose) polymerase (PARP) mechanisms of action and rationale for targeting in cancer and other diseases. Crit Rev Eukaryot Gene Expr 2014;24:15-28. [Crossref] [PubMed]
- O'Sullivan CC, Moon DH, Kohn EC, et al. Beyond Breast and Ovarian Cancers: PARP Inhibitors for BRCA Mutation-Associated and BRCA-Like Solid Tumors. Front Oncol 2014;4:42. [PubMed]
- Farmer H, McCabe N, Lord CJ, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 2005;434:917-21. [Crossref] [PubMed]
- Bryant HE, Schultz N, Thomas HD, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 2005;434:913-7. [Crossref] [PubMed]
- Scott CL, Swisher EM, Kaufmann SH. Poly (ADP-ribose) polymerase inhibitors: recent advances and future development. J Clin Oncol 2015;33:1397-406. [Crossref] [PubMed]
- Gottipati P, Vischioni B, Schultz N, et al. Poly(ADP-ribose) polymerase is hyperactivated in homologous recombination-defective cells. Cancer Res 2010;70:5389-98. [Crossref] [PubMed]
- Lakshmipathy U, Campbell C. The human DNA ligase III gene encodes nuclear and mitochondrial proteins. Mol Cell Biol 1999;19:3869-76. [Crossref] [PubMed]
- Wang H, Rosidi B, Perrault R, et al. DNA ligase III as a candidate component of backup pathways of nonhomologous end joining. Cancer Res 2005;65:4020-30. [Crossref] [PubMed]
- Akbari M, Keijzers G, Maynard S, et al. Overexpression of DNA ligase III in mitochondria protects cells against oxidative stress and improves mitochondrial DNA base excision repair. DNA Repair (Amst) 2014;16:44-53. [Crossref] [PubMed]
- Chen X, Zhong S, Zhu X, et al. Rational design of human DNA ligase inhibitors that target cellular DNA replication and repair. Cancer Res 2008;68:3169-77. [Crossref] [PubMed]
- Sallmyr A, Matsumoto Y, Roginskaya V, et al. Inhibiting Mitochondrial DNA Ligase IIIalpha Activates Caspase 1-Dependent Apoptosis in Cancer Cells. Cancer Res 2016;76:5431-41. [Crossref] [PubMed]
- Tobin LA, Robert C, Nagaria P, et al. Targeting abnormal DNA repair in therapy-resistant breast cancers. Mol Cancer Res 2012;10:96-107. [Crossref] [PubMed]
- Newman EA, Lu F, Bashllari D, et al. Alternative NHEJ Pathway Components Are Therapeutic Targets in High-Risk Neuroblastoma. Mol Cancer Res 2015;13:470-82. [Crossref] [PubMed]
- Newman EA, Chukkapalli S, Bashllari D, et al. Alternative NHEJ pathway proteins as components of MYCN oncogenic activity in human neural crest stem cell differentiation: implications for neuroblastoma initiation. Cell Death Dis 2017;8:3208. [Crossref] [PubMed]
- Fan J, Li L, Small D, et al. Cells expressing FLT3/ITD mutations exhibit elevated repair errors generated through alternative NHEJ pathways: implications for genomic instability and therapy. Blood 2010;116:5298-305. [Crossref] [PubMed]
- Hähnel PS, Enders B, Sasca D, et al. Targeting components of the alternative NHEJ pathway sensitizes KRAS mutant leukemic cells to chemotherapy. Blood 2014;123:2355-66. [Crossref] [PubMed]
- Muvarak N, Kelley S, Robert C, et al. c-MYC Generates Repair Errors via Increased Transcription of Alternative-NHEJ Factors, LIG3 and PARP1, in Tyrosine Kinase-Activated Leukemias. Mol Cancer Res 2015;13:699-712. [Crossref] [PubMed]
- Sallmyr A, Tomkinson AE, Rassool FV. Up-regulation of WRN and DNA ligase IIIalpha in chronic myeloid leukemia: consequences for the repair of DNA double-strand breaks. Blood 2008;112:1413-23. [Crossref] [PubMed]
- Caracciolo D, Di Martino MT, Amodio N, et al. miR-22 suppresses DNA ligase III addiction in multiple myeloma. Leukemia 2019;33:487-98. [Crossref] [PubMed]
- Pawlyn C, Loehr A, Ashby C, et al. Loss of heterozygosity as a marker of homologous repair deficiency in multiple myeloma: a role for PARP inhibition? Leukemia 2018;32:1561-6. [Crossref] [PubMed]
- Meier B, Gartner A. Having a direct look: analysis of DNA damage and repair mechanisms by next generation sequencing. Exp Cell Res 2014;329:35-41. [Crossref] [PubMed]

薛冉
北京大学肿瘤医院。从事肝脏及胰腺疾病相关研究工作近10年,目前于北京市肿瘤防治研究所暨北京大学肿瘤医院消化肿瘤内科/I期临床病区工作。2009年起专注肝癌发生的分子分型及分子机理研究。博士期间围绕胰腺癌展开了相关深入研究。近年主持国家自然科学基金1项(82002461)、北大医学青年培养基金A类1项(BMU2021PY010)、美捷登青年科学家研究基金1项(MJR20211110),参与完成了多项国家自然科学基金、首都医学发展科研基金等系列研究工作。任Annals of Translational Medicine(IF:3.689)、Journal of Clinical and Translational Hepatology(IF:4.108)及《中华生物医学工程》杂志青年编委。(更新时间:2021/8/24)
(本译文仅供学术交流,实际内容请以英文原文为准。)
Cite this article as: Caracciolo D, Montesano M, Tagliaferri P, Tassone P. Alternative non-homologous end joining repair: a master regulator of genomic instability in cancer. Precis Cancer Med 2019;2:8.