
Non-coding RNA regulatory networks in mesothelioma: a narrative review of their implication in innate immune signaling pathways
Introduction
Malignant mesothelioma [reviewed in (1) and (2)] is a rapidly fatal tumour arising in mesothelium, which has mesodermal origins and covers many of the important internal organs like the lungs (pleural mesothelioma), peritoneal cavities (peritoneal mesothelioma), the sacs surrounding the heart (pericardial mesothelioma) and the testis (tunica vaginalis mesothelioma). Although mesothelioma is a rare cancer, its incidence is still rising. Since the seminal experiments of Wagner (3), exposure to asbestos has been clearly identified as a cause of mesothelioma. It is estimated that 125 million people worldwide have a history of asbestos exposure. Although the use of asbestos has been banned in several countries, there are several developing nations that continue to use asbestos (1). In addition, asbestos has a very long latency period. The duration of time between exposure to asbestos and the incidence of disease is approximately 40 years (4). This means that the incidence of mesothelioma will continue to rise in the years to come.
Frequently alterations in clinical samples [reviewed in (1) and (2)] include loss of function of tumor suppressors common to several cancers such as CDKN2A and CDKN2B, but also in alterations in the NF2/Hippo signaling pathway converging on the activation of oncogenic Yes-associated protein 1 (YAP1) transcription co-activator, and in BRCA-associated protein 1 (BAP1). Less frequent alterations include mutations in TP53, in TERT promoter and in genes involved in RNA metabolism. Interestingly, recent studies reporting frequent somatic mutations in epigenetic and splicing regulators (5,6) suggest that these alterations may represent a novel hallmark of cancer (5,6).
Several of the alterations observed in mesothelioma, as we shall discuss in this review, are overall linked to RNA metabolism and may have an effect beyond tumor cells behavior because they interfere with microenvironment.
Interestingly, since 2012 we know that 75% of the human genome is transcribed into RNAs, while only 2% of these transcripts are translated into proteins (7). Therefore, 98% of the transcripts are not translated into proteins, while the contrary is observed in bacteria. The discovery that a large part of what had been called “junk DNA,” is actively transcribed and carries out crucial functions inspired the concept of the “RNA networks” (8), where RNA are the most influencing molecules in cellular function in eukaryotes, contrary to the view of protein-centered networks, which was assumed according to the knowledge acquired in prokaryotes.
It is very likely that a vast majority of non-coding transcripts adopts complex 3D structure(s) to achieve their biological functions. These “structured” RNAs act using very diverse mechanisms including RNA-RNA, RNA-ligand, RNA-protein, RNA-DNA, and RNA-substrate interactions (9). Structured RNAs are critical components of key molecular machines in the cell, such as the spliceosome, ribosome, and telomerase, and RNA structures play important roles in the control not only of mRNA but also noncoding RNA functions (10).
RNA structures have different organization levels: the first one consists of the nucleotide sequence folding on itself via Watson–Crick base-pairing to form secondary structure elements (e.g., hairpins) and unpaired regions. In vitro studies have demonstrated that it can be modified by the amount of magnesium chloride (11). RNA structures are highly dynamic and modulated by binding to partners, which adds another degree of complexity to these structures.
A recent genome-wide RNA–RNA crosslinking study (12) using PARIS (psoralen analysis of RNA interactions and structures) identified a large number and diversity of RNA duplexes. 25% of the aligned double-strand helix-forming sequences were found to be conserved between mammals, birds and reptiles, suggesting a biological function. Indeed, the evolutionary conservation of RNA secondary structure across several species is a strong indicator of function (13).
A large amount of structured duplex RNA present and conserved in non-coding RNA (ncRNA) is an additional argument for sections that will be discussed below on duplex RNA being part of the pattern recognized by the innate immune system. Across many common human cancers, a large proportion of tumors unexpectedly express high levels of interferon (IFN)-stimulated genes (ISGs, e.g., IFT1-3, IFITM1, DDX58, IFIH1) that are typically associated with anti-viral signaling (14). However, given that cancer-associated anti-viral signaling is occurring in a sterile microenvironment, this raises questions on the nature of the endogenous RNA that activates the signaling and the extent to which it influences the multitude of effects that stromal cells exert on cancer progression and therapy response.
In this review, we shall focus on ncRNA and immune signaling pathways in mesothelioma.
We present the following article in accordance with the Narrative Review Reporting Checklist (available at http://dx.doi.org/10.21037/pcm-21-4).
Endogenous ncRNAs activating and regulating innate immunity in mesothelioma
Endogenous ncRNA can activate the innate immunity after being recognized by RNA sensors (15). RNA sensors are classified into two families based on their structural motifs, leucine-rich repeat (LRR) and DExD/Hbox helicase. Nucleic acid-sensing in cancer is associated with cytosolic DExD/Hbox helicases (RIG-I-like receptors: RLR) or endosomal leucine-rich receptors (Toll-like receptors: TLR) sensors.
Cytosolic sensors and endogenous ncRNA ligands and regulators
In the cytosol, there are two nucleic acids sensing systems: the cGAS–STING pathway for the recognition of DNA (16), and the RLRs for the recognition of RNA species (17).
In this review dedicated to ncRNA we focus on RLRs. The latter include retinoic acid-inducible gene I (RIG-I, encoded by DDX58 gene), melanoma differentiation-associated protein 5 (MDA5, encoded by IFIH1 gene) and laboratory of genetics and physiology 2 (LGP2, encoded by DHX58 gene). RIG-1 and MDA5, but not LGP2, possess a caspase activation recruitment domain (CARD) necessary for downstream signaling. Oligomerization of RIG-I and MDA5 drive their association with their common adapter mitochondrial antiviral signaling protein (MAVS) (Figure 1).
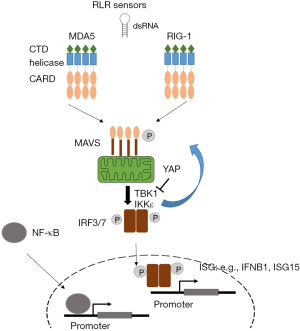
Activation of MAVS results in stimulation of the kinases TANK-binding kinase 1 (TBK1) and IκB kinase ε, which phosphorylate MAVS, then transcription factors interferon regulatory factor 3 and 7 (IRF3 and 7, respectively), and NF-κB. In the context of mesothelioma it is worth noting that YAP activity results in inhibition of TBK1 activation (18). This may represent a negative feed-back loop because dsRNA-signaling can activate YAP activity through the activation of IRF3 (19). Importantly IRF3 has been shown to act as YAP agonist and IRF3 depletion results in the suppression of YAP-driven growth (19), at least in gastric cancer. These data are consistent with our observation that dsRNA signaling is activated (20) during mesothelioma development in asbestos-exposed mice where we had previously described YAP activation (21). Altogether, this suggests that IRF3 could be a new therapeutic target against YAP-driven mesothelioma. As we mentioned above, YAP activation is controlled by the NF2/Hippo pathway but recent work has also conferred this property to BAP1 (22), at least in pancreatic cancer. Therefore, control of YAP signaling has an influence not only on tumor cell proliferation but also shuts-down a signaling affecting immune signaling. Indeed, phosphorylation of IRF3 leads to conformational changes and rearrangement of IRF3 monomers to dimers. Dimeric IRF3 then translocates to the nucleus to bind to IRF binding elements for the induction of interferon-stimulated genes (ISG), such as IFNB1, ISG15 (Figure 1) (23). As mentioned above, among the most frequent genomic alterations in pleural mesothelioma are the homozygous deletions (HD) of the CDKN2A tumor suppressor gene. CDKN2A HD are accompanied in 30% of the cases by the HD of all genes encoding type I interferons (IFN I) which lie nearby in the chromosome 9 (24), indicating an additional interference with interferon signaling.
LGP2 is unable to interact with MAVS (25) and has been found to have opposite effects on RIG-I and MDA5. Indeed, LGP2 downregulates RIG-I’s signaling activity, while it upregulates MDA5 signaling (26).
Although knowledge about these sensors had been acquired because they are essential in response to viruses, which form dsRNA during their replication, they can be stimulated by endogenously formed dsRNA, in the absence of mechanisms allowing the distinction between self and non-self nucleic acids. Indeed, RLRs utilize multiple criteria to ensure selective recognition of non-self RNA and robust discrimination against cellular RNA. For both RIG-I and MDA5, duplex RNA structure is necessary but not sufficient for the sensing of foreign RNA.
Endogenous RIG-1 ligands
For the activation of RIG-I, a 5' triphosphate group (5'ppp), which is present in all nascent transcripts and unprocessed viral RNAs, is additionally required (27-29). Polymerase III transcription is the main source of endogenous 5’ppp RNA in the absence of viral infection (30). Polymerase III activity is augmented by MYC (31) and by nearby RNA polymerase II occupancy (32).
Most cytosolic mRNAs do not have a 5’ triphosphate group because they contain both a 7-methyl guanosine cap (cap 0) and 2'-O methylation (cap 1). Cap 0 protects the molecule from 5' to 3' exonuclease cleavage and is essential for the regulation of gene expression, including splicing, nuclear export of mRNA, and translation initiation (33,34). Cap 1, is dependent on the activity of a cap1 methyltransferase 1 (CMtr1), encoded by a gene induced by type 1 IFN. 2'-O methylation is required to avoid stimulation of RIG-1. Downregulation of CMtr1 results in RIG1-dependent increased type 1 IFN activation (35). Additionally, a dsRNA length of >19 bp and blunt end is sufficient to activate RIG-1 (27).
Circular RNAs (circRNA) are recently described sequences of RNA formed by back-splicing especially in the presence of Alu repeats and complementary sequences around exons (36,37). Almost 300 circRNAs have been reported to be upregulated when mesothelioma were compared to normal mesothelial cells (38). CircRNA can stimulate RIG-I and host circRNAs normally evade recognition by RIG-I through N6-methyladenosine modification, which is recognized as endogenous RNA via N6-methyladenosine readers of YT521-B homology (YTH) family (39) (Figure 2A). Recent studies have demonstrated that inhibition of N6-methyladenosine readers increases the activity of immune checkpoint inhibitors (40). This is important since YTHDF1 is significantly enriched in sarcomatoid tumors while WTHDF2 is enriched in epithelioid tumors (41).
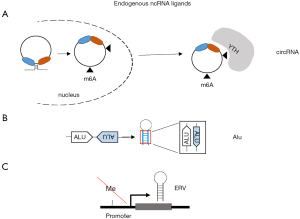
RNA processing defect leads to the accumulation of potentially immunogenic aberrant RNA transcripts. Cytoplasmic mRNA turnover is initiated by poly(A) tail removal and proceeds via two mechanisms, including 3'-5' exoribonucleolysis by the so called RNA exosome (42). Cytoplasmic RNA exosome activity requires the Ski complex, comprising the scaffold Ski3 (TTC37), two copies of Ski8 (WDR61), and the helicase Ski2 (SKIV2L). Recently, in our own experimental model of mesothelioma development (21) in mice exposed to asbestos, we observed a significant (P=7.21E-07, FDR =2.18E-06) 1.4-fold increase Skiv2l in mesothelioma tumors when compared to inflamed tissue, indicating an increase in RNA processing activity. Interestingly, SKIV2L expression is significantly higher in epithelioid compared to tissues with a sarcomatoid molecular profile (41). Serine/threonine-protein kinase/endoribonuclease inositol-requiring enzyme 1 α (IRE1α) (encoded by ERN1) is activated during the unfolded protein response (UPR), responsible for the cleavage of the precursor of XBP1 mRNA allowing the generation of the functional XBP1 necessary for the transcription of UPR genes. Interestingly, a basal UPR signaling has been observed in mesothelioma cells grown in 2D conditions and a low UPR signaling has been associated with chemotherapy resistance (43). IRE1α also generates RIG-I ligands that are normally degraded by the SKIV2L RNA exosome, thereby increasing the basal signaling activity of RIG-I in SKIV2L deficient cells (44). In the mesothelioma development model mentioned above (21) we observed a significant (P=0.00004514, FDR =0.0001064) 2-fold increase of Ern1 in mesothelioma tumors when compared to inflamed tissue, and, like for SKIV2L, ERN1 expression is significantly higher in epithelioid compared to tissues with a sarcomatoid molecular profile (41). Altogether, these observations indicate that additional ways are activated, at least in epithelial mesothelioma, to shut down interferon signaling.
Finally, depletion of the human silencing hub (HUSH) complex, which includes MPHOSPH8/MPP8, periphilin (PPHLN1) results in activation of long-interspersed nuclear elements (LINEs) repetitive elements and activation of both, RIG-I and MDA-5 (45).
MDA-5 endogenous ncRNA ligands
MDA5 does not need 5'ppp. Instead, MDA5 has more-stringent criteria for dsRNA length (>0.5–1 kb) and dsRNA complementarity of the two strands in the selective recognition of foreign dsRNA and discrimination against shorter and imperfect cellular dsRNAs. dsRNA may be present in transcripts containing repetitive elements (46) (Figure 2B). Alu are part of repetitive elements found in the human genome. There are four main types of repetitive elements: LINEs, short-interspersed nuclear elements (SINEs), Retrovirus-like elements such as endogenous retroviruses (ERV) and DNA transposon fossils (47). Altogether they cover two-third of the human genome (48). Taken together with the information that 75% of the genome is transcribed, a large part of the transcriptome corresponds to repetitive elements. Alu elements are the most important subgroup of the SINEs. They makeup 11% of the human genome and can be found in two-strand directions + and –, which allows them to bind the complementary sequence and form dsRNA structures (Figure 2B). Endogenous dsRNA formed by inverted Alu repeats, in the absence of viral infection constitute about 67% of dsRNA bound to MDA5 with a gain of function mutation G495R (49,50). Eighty-four percent of these Alus are in the 3’untranscribed region (UTR). MDA5 G495R is representative of mutations in MDA5 leading to aberrant activation of its signaling activity, resulting in a spectrum of immune disorders, such as systemic lupus erythematosus, or Aicardi-Goutières syndrome (51). In vitro, these MDA5 variants display more efficient filament formation on dsRNA and high basal signaling activities in the absence of viral infection due to misrecognition of cellular RNAs, resulting in self-triggered signaling (49). Adenosine-to-inosine modification by the adenosine deaminase acting on dsRNA (ADAR) has been found to block the recognition of dsRNA by MDA5. A defect in ADAR1 results in aberrant activation of MDA5 by cellular dsRNAs formed by Alu retroelements (49,52-54). We recently observed in an experimental animal model of asbestos-induced mesothelioma development (21), that asbestos increased the levels of RNA mutations and the most abundant changes were A to G mutations resulting from ADAR activity (55).
Aberrant activation of MDA5 is also observed due to increased synthesis of ERV (Figure 2C) upon suppression of DNA methyltransferases (56,57). Interestingly, YAP induces Dnmt3l (58) which although being inactive, stimulates the activity of the others DNMT and is important for the maintenance of embryonic stem cells. Recently, in our own model of mesothelioma development (21) we observed a significant (P=0.0008227, FDR=0.001586) 6.5-fold increase of Dnmt3l expression in mesothelioma tumors, consistent with activation of YAP signaling, when compared to inflamed tissue. In the same experimental model Dnmt1 and Dnmt3b are also significantly upregulated, consistent with the observation of methylation of some tumor suppressor genes at early stages during mesothelioma development as documented by increased levels of DNA methylation at ink4a locus after mice exposure to asbestos (59). On the other hand, Dnmt3a is significantly downregulated in both, mice (21) and rats (60), exposed to asbestos. Since Dnmt3a is the enzyme responsible for retroviral silencing in somatic cells (61), this may mean that retroviral elements may not be efficiently silenced.
Increased synthesis of ERV is also observed upon suppression of the histone methyltransferase SETDB1 (62) and this may occur in the subset of mesothelioma patients with mutated SETDB1 (41,63-65). Increased ERV expression is also observed by deficiency of the histone demethylase KDM1A/LSD1 activity (66).
The immunostimulatory effect observed by hypomethylating agents such as decitabine and azacytidine is currently exploited in combination immunotherapy treatment in different cancers (67) but not yet in mesothelioma although therapeutic approaches exploiting type-I IFN pathway signaling have already been implemented in the clinic (68) or proposed on the basis of preclinical studies (69,70).
Of particular interest in the field of cancer is that increased expression of ERV is also observed with CDK4/6 inhibitors (71). Activation of interferon signaling is observed upon inhibition of CDK4/6 in mesothelioma cells (72). This is important since CDK4/6 inhibitor abemaciclib is currently tested in p16INK4A negative MPM patients [NCT03654833 (MiST) (73).
Endosomal sensors of dsRNA
Endosomal sensing of nucleic acids is based on TLRs (74). TLR3 is a sensor for double-stranded [ds] RNA) (75), while TLR7 (76) and TLR8 are sensors for (single-stranded [ss] RNA) (77) (Figure 3). Recently, in our own model of mesothelioma development, we observed a significant increase of Tlr3, Tlr7 and Tlr8 upon asbestos exposure (21).
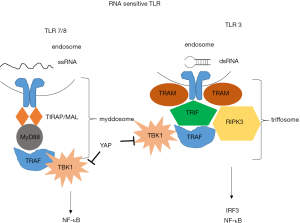
Ligand-binding mediates TLR dimerization, leading to the assembly of signaling complexes activating kinases that drive transcription and glycolysis (78). TLR7/8 induce the assembly of myddosome, upon ssRNA detection. Myddosome assembly occurs around the cytosolic tail of dimerized TLRs present at the plasma membrane or endosomes. The E3 ubiquitin ligase TNF receptor-associated factor 6 (TRAF6) is present in the myddosome. TRAF6 functions are to stimulate myddosome-associated TBK1 to drive metabolic changes in the cell and to stimulate IKK- and MAPK-dependent transcription factors. TLR3 induces the assembly of triffosome that are so called because they contain TIR domain-containing adaptor inducing IFN-β (TRIF). TRIF promotes TBK1-dependent gene expression and receptor-interacting serine/threonine-protein kinase 3-dependent necroptosis. Nucleic acids have to be processed to nucleic acids and free nucleosides in endosomes to be able to activate downstream signaling. TLR7 dimerization is most efficient in the presence of ssRNA and free guanosine molecules (79,80). Similarly, TLR8 dimers form most efficiently in the presence of ssRNA and free uridines (77). The process by which dual ligands are produced to maximally dimerize TLRs may occur only in endosomes through the actions of acid-dependent nucleases present in these organelles. Cells lacking the lysosomal RNase T2 are defective for TLR8 signaling (77). Additionally, at least in the case of TLR3, the affinity of the sensor for nucleic acids is strongest at acidic pH (81). Single molecules of dsRNA bind TLR3 dimers (82). TLR3 minimally responds to approximately 40 bp dsRNA by a reporter assay (83,84). Experimental evidence suggests activation of type 1 IFN via TLR3 by antracycline treatment (85), although the species activating the signaling is unknown. It has been reported that addition of dsRNA binding domain agonist poly (I-C) synthetic ligand induces cell death in some TLR3-positive mesothelioma cells and the effects are increased by cisplatin pre-treatment in p53 wild-type cells (69). This is consistent with the knowledge about TLR3 induction by p53 (86) .
Perspective and open questions
Knowledge recently acquired and discussed in this review highlights the role of endogenous ncRNA as ligands in innate immunity in mesothelioma. Mesothelioma is the sixth of 31 cancer types with most prevalent 38- ISGs signature (87) and, in a large fraction of ISGs high tumors, no immune cells, possibly contributing to the phenotype, have been detected, indicating spontaneous IFN production by cancer cells per se. This is consistent with recent studies showing that mesothelioma cells maintain the activation of the type-I IFN signaling pathway (20,88). Oncoprint analysis (www.cBioportal.org) of mesothelioma TCGA data (Figure 4) of the different components of the ncRNA regulatory networks involved in innate immunity described in this review shows that patients with an activated type 1 interferon signaling (increased expression of ISG) have a tendency to better overall survival. Alterations in ncRNA regulatory networks are frequently observed and there is a statistically significant co-occurrence between alterations of several components of ncRNA regulatory networks (Table 1).
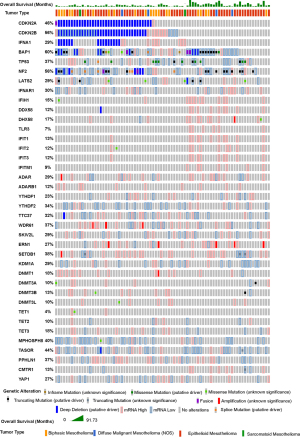
Table 1
| A | B | Neither | A Not B | B Not A | Both | Log2 Odds Ratio | p-Value | q-Value | Tendency |
|---|---|---|---|---|---|---|---|---|---|
| CDKN2A | CDKN2B | 35 | 1 | 9 | 37 | >3 | <0.001 | <0.001 | Co-occurrence |
| IFIT1 | IFIT3 | 71 | 1 | 0 | 10 | >3 | <0.001 | <0.001 | Co-occurrence |
| DDX58 | IFIT1 | 70 | 1 | 2 | 9 | >3 | <0.001 | <0.001 | Co-occurrence |
| DDX58 | IFIT3 | 70 | 2 | 2 | 8 | >3 | <0.001 | <0.001 | Co-occurrence |
| IFIT2 | IFIT3 | 70 | 2 | 2 | 8 | >3 | <0.001 | <0.001 | Co-occurrence |
| IFIT1 | IFIT2 | 69 | 3 | 2 | 8 | >3 | <0.001 | <0.001 | Co-occurrence |
| IFIH1 | IFIT3 | 67 | 5 | 3 | 7 | >3 | <0.001 | 0.002 | Co-occurrence |
| IFIH1 | IFIT1 | 66 | 5 | 4 | 7 | >3 | <0.001 | 0.005 | Co-occurrence |
| DDX58 | IFIT2 | 68 | 4 | 4 | 6 | >3 | <0.001 | 0.008 | Co-occurrence |
| CDKN2A | IFNA1 | 39 | 19 | 5 | 19 | 2.963 | <0.001 | 0.009 | Co-occurrence |
| IFIH1 | TLR3 | 69 | 7 | 1 | 5 | >3 | <0.001 | 0.009 | Co-occurrence |
| CDKN2B | IFNA1 | 33 | 25 | 3 | 21 | >3 | <0.001 | 0.009 | Co-occurrence |
| IFIT3 | IFITM1 | 70 | 5 | 2 | 5 | >3 | <0.001 | 0.009 | Co-occurrence |
| DDX58 | CMTR1 | 67 | 4 | 5 | 6 | >3 | <0.001 | 0.011 | Co-occurrence |
| IFIT1 | IFITM1 | 69 | 6 | 2 | 5 | >3 | <0.001 | 0.015 | Co-occurrence |
| TLR3 | ADAR | 58 | 0 | 18 | 6 | >3 | <0.001 | 0.016 | Co-occurrence |
| IFIH1 | DDX58 | 66 | 6 | 4 | 6 | >3 | <0.001 | 0.016 | Co-occurrence |
| IFIH1 | IFIT2 | 66 | 6 | 4 | 6 | >3 | <0.001 | 0.016 | Co-occurrence |
| IFIT1 | CMTR1 | 66 | 5 | 5 | 6 | >3 | <0.001 | 0.017 | Co-occurrence |
| CDKN2A | MPHOSPH8 | 34 | 15 | 10 | 23 | 2.382 | <0.001 | 0.017 | Co-occurrence |
| IFIH1 | IFITM1 | 68 | 7 | 2 | 5 | >3 | <0.001 | 0.017 | Co-occurrence |
| NF2 | IFNAR1 | 32 | 25 | 4 | 21 | 2.748 | <0.001 | 0.019 | Co-occurrence |
| DNMT3A | TET1 | 74 | 5 | 0 | 3 | >3 | <0.001 | 0.019 | Co-occurrence |
| CDKN2B | IFIT1 | 26 | 45 | 10 | 1 | <-3 | <0.001 | 0.026 | Mutual exclusivity |
| TP53 | DNMT3B | 50 | 21 | 2 | 9 | >3 | 0.001 | 0.040 | Co-occurrence |
| DDX58 | TLR3 | 70 | 6 | 2 | 4 | >3 | 0.002 | 0.040 | Co-occurrence |
| TLR3 | IFIT2 | 70 | 2 | 6 | 4 | >3 | 0.002 | 0.040 | Co-occurrence |
| TLR3 | IFIT3 | 70 | 2 | 6 | 4 | >3 | 0.002 | 0.040 | Co-occurrence |
| CDKN2B | DDX58 | 27 | 45 | 9 | 1 | <-3 | 0.002 | 0.049 | Mutual exclusivity |
| CDKN2B | IFIT3 | 27 | 45 | 9 | 1 | <-3 | 0.002 | 0.049 | Mutual exclusivity |
| CDKN2B | IFITM1 | 29 | 46 | 7 | 0 | <-3 | 0.002 | 0.049 | Mutual exclusivity |
| TTC37 | TET3 | 51 | 16 | 5 | 10 | 2.672 | 0.002 | 0.049 | Co-occurrence |
| TLR3 | IFIT1 | 69 | 2 | 7 | 4 | >3 | 0.002 | 0.049 | Co-occurrence |
| TLR3 | CMTR1 | 69 | 2 | 7 | 4 | >3 | 0.002 | 0.049 | Co-occurrence |
| DHX58 | IFIT1 | 63 | 8 | 5 | 6 | >3 | 0.002 | 0.049 | Co-occurrence |
| CDKN2A | CDKN2B | 35 | 1 | 9 | 37 | >3 | <0.001 | <0.001 | Co-occurrence |
| IFIT1 | IFIT3 | 71 | 1 | 0 | 10 | >3 | <0.001 | <0.001 | Co-occurrence |
| DDX58 | IFIT1 | 70 | 1 | 2 | 9 | >3 | <0.001 | <0.001 | Co-occurrence |
| DDX58 | IFIT3 | 70 | 2 | 2 | 8 | >3 | <0.001 | <0.001 | Co-occurrence |
The more detailed mechanisms behind the crosstalk between cancer therapy and stimulation of innate immune system by endogenous ncRNA characterized so far are related to irradiation or inhibition of DNA methyltransferase, which are already explored in clinical trials. Few clinical trials have or are exploring synthetic RNA as direct stimulators (89,90). For example, pre-clinical studies have shown that modified poly-IC, a synthetic mimic of dsRNA, increases tumor infiltration by T cells (91) by stimulating MDA5-mediated production of IFN β in vascular endothelial cells, suggesting that this stimulation may improve T-cell based cancer immunotherapy.
Besides being used for therapeutic intervention, the expression of endogenous ncRNA could be used as in mesothelioma as biomarkers e.g., to stratify patients responding to immunotherapy as it has been shown for ERV in clear cell kidney cancer (92).
On the other side cancer cells impairment of type 1 IFN signaling makes them sensitive to oncolytic therapy (93), indicating ways to select the patients.
While several experimental mechanistic data and clinical evidence (94) have highlighted the contribution of endogenous ncRNA for cancer patients to obtain clinical benefits from immunogenic cell death-inducing therapies, several questions still subsist. This includes the extent of the variation of endogenous ncRNA ligands between individuals. Advances in sequencing techniques and global determination of RNA structures in living cells, such as the PARIS technique mentioned in the introduction, will allow the analysis of the functions of RNA structures, therefore laying a foundation for understanding RNA regulatory networks and immune signaling pathways in mesothelioma. However, as previously mentioned ncRNA could already be explored as potential markers for therapeutic intervention such as ERV for the stratification of patients for immunotherapy.
Acknowledgments
Funding: This work was supported by Swiss National Science Foundation grant 320030_182690, Walter-Bruckerhoff Stiftung and Stiftung für Angewandte Krebsforchung.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editor (Mauro Tognon) for the series “Malignant Pleural Mesothelioma” published in Precision Cancer Medicine. The article has undergone external peer review.
Reporting Checklist: The author has completed the Narrative Review Reporting Checklist. Available at http://dx.doi.org/10.21037/pcm-21-4
Conflicts of Interest: The author has completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/pcm-21-4). The series “Malignant Pleural Mesothelioma” was commissioned by the editorial office without any funding or sponsorship. The author has no other conflicts of interest to declare.
Ethical Statement: The author is accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Felley-Bosco E, MacFarlane M. Asbestos: Modern Insights for Toxicology in the Era of Engineered Nanomaterials. Chem Res Toxicol 2018;31:994-1008. [Crossref] [PubMed]
- Carbone M, Adusumilli PS, Alexander HR Jr, et al. Mesothelioma: Scientific clues for prevention, diagnosis, and therapy. CA Cancer J Clin 2019;69:402-29. [Crossref] [PubMed]
- Wagner JC. Experimental production of mesothelial tumours of the pleura by implantation of dusts in laboratory animals. Nature 1962;196:180-1. [Crossref] [PubMed]
- van Meerbeeck JP, Scherpereel A, Surmont VF, et al. Malignant pleural mesothelioma: The standard of care and challenges for future management. Crit Rev Oncol Hematol 2011;78:92-111. [Crossref] [PubMed]
- Elsässer SJ, Allis CD, Lewis PW. Cancer. New epigenetic drivers of cancers. Science 2011;331:1145-6. [Crossref] [PubMed]
- Imielinski M, Berger AH, Hammerman PS, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell 2012;150:1107-20. [Crossref] [PubMed]
- ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 2012;489:57-74. [Crossref] [PubMed]
- Vandevenne M, Delmarcelle M, Galleni M. RNA Regulatory Networks as a Control of Stochasticity in Biological Systems. Front Genet 2019;10:403. [Crossref] [PubMed]
- Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell 2011;43:904-14. [Crossref] [PubMed]
- Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell 2014;157:77-94. [Crossref] [PubMed]
- Mu X, Greenwald E, Ahmad S, et al. An origin of the immunogenicity of in vitro transcribed RNA. Nucleic Acids Res 2018;46:5239-49. [Crossref] [PubMed]
- Lu Z, Zhang QC, Lee B, et al. RNA Duplex Map in Living Cells Reveals Higher-Order Transcriptome Structure. Cell 2016;165:1267-79. [Crossref] [PubMed]
- Smith MA, Gesell T, Stadler PF, et al. Widespread purifying selection on RNA structure in mammals. Nucleic Acids Res 2013;41:8220-36. [Crossref] [PubMed]
- Weichselbaum RR, Ishwaran H, Yoon T, et al. An interferon-related gene signature for DNA damage resistance is a predictive marker for chemotherapy and radiation for breast cancer. Proc Natl Acad Sci U S A 2008;105:18490-5. [Crossref] [PubMed]
- Bartok E, Hartmann G. Immune Sensing Mechanisms that Discriminate Self from Altered Self and Foreign Nucleic Acids. Immunity 2020;53:54-77. [Crossref] [PubMed]
- Ablasser A, Chen ZJ. cGAS in action: Expanding roles in immunity and inflammation. Science 2019;363:eaat8657. [Crossref] [PubMed]
- Hur S. Double-Stranded RNA Sensors and Modulators in Innate Immunity. Annu Rev Immunol 2019;37:349-75. [Crossref] [PubMed]
- Zhang Q, Meng F, Chen S, et al. Hippo signalling governs cytosolic nucleic acid sensing through YAP/TAZ-mediated TBK1 blockade. Nat Cell Biol 2017;19:362-74. [Crossref] [PubMed]
- Jiao S, Guan J, Chen M, et al. Targeting IRF3 as a YAP agonist therapy against gastric cancer. J Exp Med 2018;215:699-718. [Crossref] [PubMed]
- Sun S, Frontini F, Qi W, et al. Endogenous retrovirus expression activates type-I interferon signaling in an experimental mouse model of mesothelioma development. Cancer Lett 2021;507:26-38. [Crossref] [PubMed]
- Rehrauer H, Wu L, Blum W, et al. How asbestos drives the tissue towards tumors: YAP activation, macrophage and mesothelial precursor recruitment, RNA editing, and somatic mutations. Oncogene 2018;37:2645-59. [Crossref] [PubMed]
- Lee HJ, Pham T, Chang MT, et al. The Tumor Suppressor BAP1 Regulates the Hippo Pathway in Pancreatic Ductal Adenocarcinoma. Cancer Res 2020;80:1656-68. [Crossref] [PubMed]
- Liu S, Cai X, Wu J, et al. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science 2015;347:aaa2630. [Crossref] [PubMed]
- Delaunay T, Achard C, Boisgerault N, et al. Frequent Homozygous Deletions of Type I Interferon Genes in Pleural Mesothelioma Confer Sensitivity to Oncolytic Measles Virus. J Thorac Oncol 2020;15:827-42. [Crossref] [PubMed]
- Yoneyama M, Kikuchi M, Matsumoto K, et al. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J Immunol 2005;175:2851-8. [Crossref] [PubMed]
- Ablasser A, Hur S. Regulation of cGAS- and RLR-mediated immunity to nucleic acids. Nat Immunol 2020;21:17-29. [Crossref] [PubMed]
- Schlee M. Master sensors of pathogenic RNA - RIG-I like receptors. Immunobiology 2013;218:1322-35. [Crossref] [PubMed]
- Pichlmair A, Schulz O, Tan CP, et al. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5'-phosphates. Science 2006;314:997-1001. [Crossref] [PubMed]
- Hornung V, Ellegast J, Kim S, et al. 5'-Triphosphate RNA is the ligand for RIG- I. Science 2006;314:994-7. [Crossref] [PubMed]
- White RJ. Transcription by RNA polymerase III: more complex than we thought. Nat Rev Genet 2011;12:459-63. [Crossref] [PubMed]
- Gomez-Roman N, Grandori C, Eisenman RN, et al. Direct activation of RNA polymerase III transcription by c-Myc. Nature 2003;421:290-4. [Crossref] [PubMed]
- Oler AJ, Alla RK, Roberts DN, et al. Human RNA polymerase III transcriptomes and relationships to Pol II promoter chromatin and enhancer-binding factors. Nat Struct Mol Biol 2010;17:620-8. [Crossref] [PubMed]
- Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell 2009;136:688-700. [Crossref] [PubMed]
- Cowling VH. Regulation of mRNA cap methylation. Biochem J 2009;425:295-302. [Crossref] [PubMed]
- Schuberth-Wagner C, Ludwig J, Bruder AK, et al. A Conserved Histidine in the RNA Sensor RIG-I Controls Immune Tolerance to N1-2'O-Methylated Self RNA. Immunity 2015;43:41-51. [Crossref] [PubMed]
- Zhang XO, Wang HB, Zhang Y, et al. Complementary sequence-mediated exon circularization. Cell 2014;159:134-47. [Crossref] [PubMed]
- Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013;19:141-57. [Crossref] [PubMed]
- Winata PPW, Cheng YYC, Williams MMW, et al. Circular RNAs as novel biomarkers for detection of malignant pleural mesothelioma. Asia-Pacific Journal of Clinical Oncology 2018;14:82-3.
- Chen YG, Chen R, Ahmad S, et al. N6-Methyladenosine Modification Controls Circular RNA Immunity. Mol Cell 2019;76:96-109.e9. [Crossref] [PubMed]
- Han D, Liu J, Chen C, et al. Anti-tumour immunity controlled through mRNA m(6)A methylation and YTHDF1 in dendritic cells. Nature 2019;566:270-4. [Crossref] [PubMed]
- Bueno R, Stawiski EW, Goldstein LD, et al. Comprehensive genomic analysis of malignant pleural mesothelioma identifies recurrent mutations, gene fusions and splicing alterations. Nat Genet 2016;48:407-16. [Crossref] [PubMed]
- Łabno A, Tomecki R, Dziembowski A. Cytoplasmic RNA decay pathways - Enzymes and mechanisms. Biochim Biophys Acta 2016;1863:3125-47. [Crossref] [PubMed]
- Xu D, Liang SQ, Yang H, et al. Increased sensitivity to apoptosis upon endoplasmic reticulum stress-induced activation of the unfolded protein response in chemotherapy-resistant malignant pleural mesothelioma. Br J Cancer 2018;119:65-75. [Crossref] [PubMed]
- Eckard SC, Rice GI, Fabre A, et al. The SKIV2L RNA exosome limits activation of the RIG-I-like receptors. Nat Immunol 2014;15:839-45. [Crossref] [PubMed]
- Tunbak H, Enriquez-Gasca R, Tie CHC, et al. The HUSH complex is a gatekeeper of type I interferon through epigenetic regulation of LINE-1s. Nat Commun 2020;11:5387. [Crossref] [PubMed]
- Deininger P. Alu elements: know the SINEs. Genome Biol 2011;12:236. [Crossref] [PubMed]
- Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature 2001;409:860-921. Erratum in: Nature 2001;412:565 Erratum in: Nature 2001;411:720. [Crossref] [PubMed]
- de Koning AP, Gu W, Castoe TA, et al. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet 2011;7:e1002384. [Crossref] [PubMed]
- Ahmad S, Mu X, Yang F, et al. Breaching Self-Tolerance to Alu Duplex RNA Underlies MDA5-Mediated Inflammation. Cell 2018;172:797-810 e13. [Crossref] [PubMed]
- Kato H, Takeuchi O, Mikamo-Satoh E, et al. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med 2008;205:1601-10. [Crossref] [PubMed]
- Crow YJ, Manel N. Aicardi-Goutieres syndrome and the type I interferonopathies. Nat Rev Immunol 2015;15:429-40. [Crossref] [PubMed]
- Mannion NM, Greenwood SM, Young R, et al. The RNA-editing enzyme ADAR1 controls innate immune responses to RNA. Cell Rep 2014;9:1482-94. [Crossref] [PubMed]
- Liddicoat BJ, Piskol R, Chalk AM, et al. RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself. Science 2015;349:1115-20. [Crossref] [PubMed]
- Pestal K, Funk CC, Snyder JM, et al. Isoforms of RNA-Editing Enzyme ADAR1 Independently Control Nucleic Acid Sensor MDA5-Driven Autoimmunity and Multi-organ Development. Immunity 2015;43:933-44. [Crossref] [PubMed]
- Nishikura K. A-to-I editing of coding and non-coding RNAs by ADARs. Nat Rev Mol Cell Biol 2016;17:83-96. [Crossref] [PubMed]
- Chiappinelli KB, Strissel PL, Desrichard A, et al. Inhibiting DNA Methylation Causes an Interferon Response in Cancer via dsRNA Including Endogenous Retroviruses. Cell 2015;162:974-86. Erratum in: Cell 2016;164:1073 Buhu, Sadna [corrected to Budhu, Sadna]; Mergoub, Taha [corrected to Merghoub, Taha]. Erratum in: Cell 2017;169:361. [Crossref] [PubMed]
- Roulois D, Loo Yau H, Singhania R, et al. DNA-Demethylating Agents Target Colorectal Cancer Cells by Inducing Viral Mimicry by Endogenous Transcripts. Cell 2015;162:961-73. [Crossref] [PubMed]
- Lian I, Kim J, Okazawa H, et al. The role of YAP transcription coactivator in regulating stem cell self-renewal and differentiation. Genes Dev 2010;24:1106-18. [Crossref] [PubMed]
- Chernova T, Murphy FA, Galavotti S, et al. Long-Fiber Carbon Nanotubes Replicate Asbestos-Induced Mesothelioma with Disruption of the Tumor Suppressor Gene Cdkn2a (Ink4a/Arf). Curr Biol 2017;27:3302-14.e6. [Crossref] [PubMed]
- Roulois D, Deshayes S, Guilly MN, et al. Characterization of preneoplastic and neoplastic rat mesothelial cell lines: the involvement of TETs, DNMTs, and 5-hydroxymethylcytosine. Oncotarget 2016;7:34664-87. [Crossref] [PubMed]
- Kao TH, Liao HF, Wolf D, et al. Ectopic DNMT3L triggers assembly of a repressive complex for retroviral silencing in somatic cells. J Virol 2014;88:10680-95. [Crossref] [PubMed]
- Cuellar TL, Herzner AM, Zhang X, et al. Silencing of retrotransposons by SETDB1 inhibits the interferon response in acute myeloid leukemia. J Cell Biol 2017;216:3535-49. [Crossref] [PubMed]
- Kang HC, Kim HK, Lee S, et al. Whole exome and targeted deep sequencing identify genome-wide allelic loss and frequent SETDB1 mutations in malignant pleural mesotheliomas. Oncotarget 2016;7:8321-31. [Crossref] [PubMed]
- Hmeljak J, Sanchez-Vega F, Hoadley KA, et al. Integrative Molecular Characterization of Malignant Pleural Mesothelioma. Cancer Discov 2018;8:1548-65. [Crossref] [PubMed]
- Pagano M, Ceresoli LG, Zucali PA, et al. Mutational Profile of Malignant Pleural Mesothelioma (MPM) in the Phase II RAMES Study. Cancers (Basel) 2020;12:2948. [Crossref] [PubMed]
- Sheng W, LaFleur MW, Nguyen TH, et al. LSD1 Ablation Stimulates Anti-tumor Immunity and Enables Checkpoint Blockade. Cell 2018;174:549-63.e19. [Crossref] [PubMed]
- Mehdipour P, Murphy T, De Carvalho DD. The role of DNA-demethylating agents in cancer therapy. Pharmacol Ther 2020;205:107416. [Crossref] [PubMed]
- Sterman DH, Recio A, Haas AR, et al. A phase I trial of repeated intrapleural adenoviral-mediated interferon-beta gene transfer for mesothelioma and metastatic pleural effusions. Mol Ther 2010;18:852-60. [Crossref] [PubMed]
- Vanbervliet-Defrance B, Delaunay T, Daunizeau T, et al. Cisplatin unleashes Toll-like receptor 3-mediated apoptosis through the downregulation of c-FLIP in malignant mesothelioma. Cancer Lett 2020;472:29-39. [Crossref] [PubMed]
- Achard C, Boisgerault N, Delaunay T, et al. Sensitivity of human pleural mesothelioma to oncolytic measles virus depends on defects of the type I interferon response. Oncotarget 2015;6:44892-904. [Crossref] [PubMed]
- Goel S, DeCristo MJ, Watt AC, et al. CDK4/6 inhibition triggers anti-tumour immunity. Nature 2017;548:471-5. [Crossref] [PubMed]
- Aliagas E, Martinez-Iniesta M, Hernandez M, et al. CDK4/6 Inhibitors Show Antitumor Effects in 1. Preclinical Models of Malignant Pleural Mesothelioma. J Thorac Oncol 2019;14:S343. [Crossref]
- Cantini L, Hassan R, Sterman DH, et al. Emerging Treatments for Malignant Pleural Mesothelioma: Where Are We Heading? Front Oncol 2020;10:343. [Crossref] [PubMed]
- Janeway CA Jr, Medzhitov R. Innate immune recognition. Annu Rev Immunol 2002;20:197-216. [Crossref] [PubMed]
- Alexopoulou L, Holt AC, Medzhitov R, et al. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature 2001;413:732-8. [Crossref] [PubMed]
- Diebold SS, Kaisho T, Hemmi H, et al. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science 2004;303:1529-31. [Crossref] [PubMed]
- Greulich W, Wagner M, Gaidt MM, et al. TLR8 Is a Sensor of RNase T2 Degradation Products. Cell 2019;179:1264-75.e13. [Crossref] [PubMed]
- Fitzgerald KA, Kagan JC. Toll-like Receptors and the Control of Immunity. Cell 2020;180:1044-66. [Crossref] [PubMed]
- Shibata T, Ohto U, Nomura S, et al. Guanosine and its modified derivatives are endogenous ligands for TLR7. Int Immunol 2016;28:211-22. [Crossref] [PubMed]
- Zhang Z, Ohto U, Shibata T, et al. Structural Analysis Reveals that Toll-like Receptor 7 Is a Dual Receptor for Guanosine and Single-Stranded RNA. Immunity 2016;45:737-48. [Crossref] [PubMed]
- de Bouteiller O, Merck E, Hasan UA, et al. Recognition of double-stranded RNA by human toll-like receptor 3 and downstream receptor signaling requires multimerization and an acidic pH. J Biol Chem 2005;280:38133-45. [Crossref] [PubMed]
- Choe J, Kelker MS, Wilson IA. Crystal structure of human toll-like receptor 3 (TLR3) ectodomain. Science 2005;309:581-5. [Crossref] [PubMed]
- Tatematsu M, Nishikawa F, Seya T, et al. Toll-like receptor 3 recognizes incomplete stem structures in single-stranded viral RNA. Nat Commun 2013;4:1833. [Crossref] [PubMed]
- Jelinek I, Leonard JN, Price GE, et al. TLR3-specific double-stranded RNA oligonucleotide adjuvants induce dendritic cell cross-presentation, CTL responses, and antiviral protection. J Immunol 2011;186:2422-9. [Crossref] [PubMed]
- Sistigu A, Yamazaki T, Vacchelli E, et al. Cancer cell-autonomous contribution of type I interferon signaling to the efficacy of chemotherapy. Nat Med 2014;20:1301-9. [Crossref] [PubMed]
- Shatz M, Menendez D, Resnick MA. The human TLR innate immune gene family is differentially influenced by DNA stress and p53 status in cancer cells. Cancer Res 2012;72:3948-57. [Crossref] [PubMed]
- Liu H, Golji J, Brodeur LK, et al. Tumor-derived IFN triggers chronic pathway agonism and sensitivity to ADAR loss. Nat Med 2019;25:95-102. [Crossref] [PubMed]
- Chernova T, Sun XM, Powley IR, et al. Molecular profiling reveals primary mesothelioma cell lines recapitulate human disease. Cell Death Differ 2016;23:1152-64. [Crossref] [PubMed]
- Frega G, Wu Q, Le Naour J, et al. Trial Watch: experimental TLR7/TLR8 agonists for oncological indications. Oncoimmunology 2020;9:1796002. [Crossref] [PubMed]
- Le Naour J, Galluzzi L, Zitvogel L, et al. Trial watch: TLR3 agonists in cancer therapy. Oncoimmunology 2020;9:1771143. [Crossref] [PubMed]
- Sultan H, Wu J, Fesenkova VI, et al. Poly-IC enhances the effectiveness of cancer immunotherapy by promoting T cell tumor infiltration. J Immunother Cancer 2020;8:e001224. [Crossref] [PubMed]
- Smith CC, Beckermann KE, Bortone DS, et al. Endogenous retroviral signatures predict immunotherapy response in clear cell renal cell carcinoma. J Clin Invest 2018;128:4804-20. [Crossref] [PubMed]
- Matveeva OV, Chumakov PM. Defects in interferon pathways as potential biomarkers of sensitivity to oncolytic viruses. Rev Med Virol 2018;28:e2008. [Crossref] [PubMed]
- Galluzzi L, Vitale I, Warren S, et al. Consensus guidelines for the definition, detection and interpretation of immunogenic cell death. J Immunother Cancer 2020;8:e000337. Erratum in: J Immunother Cancer 2020;8. [Crossref] [PubMed]
Cite this article as: Felley-Bosco E. Non-coding RNA regulatory networks in mesothelioma: a narrative review of their implication in innate immune signaling pathways. Precis Cancer Med 2021;4:23.

