
Anti-proliferative and anti-migratory effects of EGFR and c-Met tyrosine kinase inhibitors in triple negative breast cancer cells
Introduction
Breast cancer is the most commonly diagnosed neoplasm in women, with over 279,000 American cases expected to be diagnosed in 2020 (1). Although mortality associated with breast cancer has declined due to advancements in screening and therapeutics, it remains a lethal disease with a projected rate of 22 deaths per 100,000 in 2020 (2). Breast cancer is a heterogenous disease and prognosis is dependent on tumor expression of the receptors estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2/neu) as they present effective therapeutic targets. Based on this classification, breast cancer is largely divided into four clinical subtypes: Luminal A (ER/PR+, low ki67, usually HER2−), luminal B (ER/PR+, high ki67, HER2+/−), HER2-enriched (HER2+, ER/PR−) and triple negative breast cancer (TNBC) (ER/PR−, HER2−) (3,4). For most patients with breast cancer, treatment entails surgery with radiation therapy and/or systemic therapy, which, depending on subtype, includes targeting of hormone receptor and growth factor receptor pathways with anti-estrogen/progesterone (tamoxifen, aromatase inhibitors) and/or anti-HER2 therapies (Herceptin®/trastuzumab, lapatinib) (5,6). However, TNBC [approximately 15–20% of cases (7)] currently does not respond to these and other molecular-targeted therapies (8-10) and tends to have a worse prognosis and lower chance of survival compared to other breast cancer subtypes (11,12). This lack of effective systemic treatment options presents an opportunity to search for targetable receptors in TNBC in order to develop more effective therapies.
For targeted therapies, kinases—enzymes responsible for phosphorylation—have been successfully used as targets in treating cancer since development of the Bcr-Abl kinase inhibitor imatinib in treating chronic myelogenous leukemia (CML) (13). In particular, the receptor tyrosine kinase (RTK) subset of membrane-bound protein receptors is of great interest for targeting. The involvement of RTKs in cancer was first observed in the 1980s with the discovery of the v-ERBB and ERBB2/neu oncogenes (14-16). It has since been demonstrated that RTKs mediate signaling for several biological processes commonly dysregulated in tumorigenesis, including cell growth/proliferation, survival, motility and invasion (17,18). These kinases are critical in driving cancer development and progression to the point where tumors may develop a reliance on kinase-driven signaling (19). This has been successfully exploited by targeted therapies such as the anti-HER2 antibody trastuzumab in HER2-enriched breast cancer (5), the anti-EGFR inhibitors gefitinib and erlotinib in non-small cell lung cancer (NSCLC) (20,21), and imatinib in CML (13). Reliance on kinase signaling can be driven by mutation and/or overexpression of the kinase and/or its ligands (19).
TNBCs often have increased expression of RTKs such as epidermal growth factor receptor (EGFR) and the c-Met (hepatocyte growth factor receptor) (22-24). EGFR and c-Met signaling involves extensive crosstalk with each other and several redundant downstream pathways (25,26) including the PI3K/AKT (27), STAT (28), and Ras (29) pathways, thus studying the effects of inhibition on both receptors individually is of great interest. Overexpression of EGFR is associated with poor outcome in various malignancies (30-32) including breast cancer (30,33), and EGFR overexpression is more frequently found in triple negative/basal-like breast cancer compared to other molecular subtypes (34-36). Similarly, c-Met has been shown to be more highly expressed in basal-like breast cancers (29,30) and TNBCs (21-23) when compared to other breast cancer subtypes. Amplification of c-Met is prognostic for higher relapse rates in patients with TNBC, compared to patients with unamplified c-Met TNBC cases (31) and c-Met overexpression is a useful prognosis factor for recurrence and death in patients with TNBC (32,33). Dysregulation of either EGFR (37,38) or c-Met (34-36) pathways plays an important role in tumorigenesis and metastasis via their regulation of cell behaviors such as proliferation, motility, apoptotic evasion, and invasion. Given the reliance of cancer progression on these pathways, EGFR and c-Met present as potential effective targets for targeted therapies in TNBC.
Erlotinib (Tarceva®, OSI-774, R1415, CP358774, NSC718781) is a reversible small molecule ATP-competing inhibitor of EGFR. Binding to the ATP-binding pocket, erlotinib prevents trans-autophosphorylation and activation of downstream cell cycle progression, proliferation, and angiogenesis signaling pathways (39). In the breast cancer setting, erlotinib has demonstrated anti-growth effects on breast cancer tumors with positive EGFR expression (40) and limited activity in unselected patients with previously untreated metastatic breast cancer (41). More specifically, erlotinib demonstrated tumor growth inhibitory effects in a TNBC xenograft model (42) with some encouraging results in phase I and phase II clinical trials to date (40,43,44).
Cabozantinib (XL184, BMS907351) is a small molecule ATP-competitive inhibitor of c-Met and VEGFR2 that is clinically used for treatment against medullary thyroid cancer and hepatocellular carcinoma. In preclinical studies, cabozantinib has been shown to inhibit the phosphorylation and activation of c-Met and downstream signaling molecules in vitro (45) and in vivo (46). Moreover, cabozantinib inhibits cell proliferation and tumor growth of cancer in vitro and in vivo (37,46) with results from phase II clinical trials in metastatic breast carcinoma demonstrating initial clinical activity during treatment (38,39). Although erlotinib and cabozantinib may have some clinical activity in controlling disease progression at treatment onset, ultimately, tumors continue to progress and develop resistance to treatment. Therefore, further research is need to elucidate and understand the underlying mechanisms of c-Met and EGFR inhibition by cabozantinib and erlotinib, respectively.
In the current study, we explored the potential of EGFR and c-Met as potential targets for treatment of metastatic TNBC, including assessment of potential mechanisms of response. Using clinical data from The Cancer Genome Atlas (TCGA) and Clinical Proteomic Tumor Analysis Consortium (CPTAC), we observed that expression of both EGFR and c-Met is elevated in patients with TNBC relative to other breast cancer subtypes. Using metastatic human MDA-MB-468 and MDA-MB-231 TNBC cell lines as model systems, we observed that erlotinib and cabozantinib significantly inhibit the proliferation, migration, invasion and downstream signaling of TNBC in a cell line-specific manner, with MDA-MB-468 cells demonstrating response to erlotinib and MDA-MB-231 cells demonstrating response to cabozantinib. Elucidation of the underlying mechanisms that define the heterogenous response to tyrosine kinase inhibitors (TKIs) in TNBC could help identify biomarkers to stratify patients for treatment and/or facilitate discovery of targets to attenuate acquired resistance. We present the following article in accordance with the Materials Design Analysis Reporting (MDAR) Checklist (available at: http://dx.doi.org/10.21037/pcm-20-62).
Methods
TCGA and CPTAC analysis
RNA expression data for EGFR and c-Met in clinical breast cancer samples were generated from RNA-seq datasets of the TCGA-Breast Invasive Carcinoma (TCGA-BRCA) project by TCGA Research Network public database (https://www.cancer.gov/tcga). Protein expression data for EGFR and c-Met in clinical breast cancer samples were generated from the National Cancer Institute Clinical Proteomic Tumor Analysis Consortium (CPTAC). Expression data were retrieved via the UALCAN portal (47). Logarithmic values of RNA and protein expression were normalized to all samples and expressed as Z-values.
Cell culture and reagents
The MDA-MB-231(RRID:CVCL_0062) and MDA-MB-468(RRID:CVCL_0419) cell lines [obtained from Dr. Ann Chambers (London Health Science Centre, London, Canada)] were cultured under standard conditions in 5% CO2 at 37 °C in Dulbecco’s modified Eagle’s medium (DMEM) and nutrient mixture F12 1:1 (ThermoFisher, Waltham, MA, USA) with 10% fetal bovine serum (FBS; VWR Life Science, Radnor, PA, USA), and minimum essential medium Eagle-alpha modifications (α-MEM; ThermoFisher) with 10% FBS, respectively. Cells were cultured with the EGFR inhibitor erlotinib (LC Laboratories, Woburn, MA, USA) or the c-Met inhibitor cabozantinib (MedChemExpress, Monmouth Junction, NJ, USA). When assessing EGFR and c-Met signaling, cells were incubated with 100 ng/mL EGF (Sigma-Aldrich, Mississauga, ON, Canada) and 50 ng/mL HGF (Abcam, Cambridge, UK), respectively.
Proliferation assays
To assess the effects of EGFR and c-Met inhibitors on growth kinetics and cell proliferation, growth curve and clonogenic assays were performed. For growth curve assays, cells were seeded in 60 mm dishes at density of 5×104 cells/dish, allowed to attach after several hours, and then incubated with either the inhibitor (0.5–5 µM erlotinib; 1–2 µM cabozantinib) or dimethyl sulfoxide (DMSO, Sigma-Aldrich; vehicle control). Cell were incubated at 37 °C, 5% CO2 for 9 days, with media containing inhibitors/DMSO refreshed every 4 days. At days 1, 3, 5, 7, and 9 cells were harvested by trypsinization and counted with a haemocytometer using trypan blue exclusion. Doubling time (TD) was calculated using the formula: , where, where Nt is number of cells after time period, t, and No is the initial starting number of cells. For clonogenic assays, cells were seeded in 6-well plates at density of 100–150 cells/well, allowed to attach for several hours, and incubated with either inhibitor (0.01 nM–100 µM erlotinib; 1 nM–100 µM cabozantinib) or DMSO. Colonies were allowed to develop over 10–14 days at 37 °C, 5% CO2, with media containing inhibitors/DMSO refreshed every 4 days. At assay endpoint, colonies were fixed and stained with 6% glutaraldehyde +0.5% crystal violet solution. The mean number of colonies (defined as >50 cells) per well was calculated using FIJI ImageJ software (NIH, version 1.52p, Bethesda, MD, USA; RRID:SCR_002285).
Protein analysis
For protein analysis, lysis buffer (50 mM Tris-HCl, 150 mM NaCl, 1% NP-40, 1 mM EDTA, 1X protease inhibitor) was added to erlotinib-treated MDA-MB-468 or cabozantinib-treated MDA-MB-231 cells. Protein (30 µg per sample), quantified by Lowry assay, was boiled for 10 min with sodium dodecyl sulfate (SDS), subjected to SDS polyacrylamide gel electrophoresis (SDS-PAGE; 150 V for 1 h) and transferred onto polyvinylidene difluoride (PVDF) membranes (Millipore, Sigma-Aldrich). Membranes were blocked using 5% bovine serum albumin (BSA) in Tris-buffered saline (TBST) + 0.1% Tween-20. A summary of primary antibodies used can be found in Table S1. Secondary antibodies included goat anti-mouse IgG or goat anti-rabbit IgG secondary antibodies (Calbiochem, Billerica, MA, USA) conjugated to horseradish peroxidase were used at concentrations of 1:1,000, diluted in 5% BSA in TBST. Protein expression was visualized using AmershamTM ECLTM Prime Detection Reagent (GE Healthcare Lifesciences, Wauwatosa, WI, USA). When assessing erlotinib-induced EGFR attenuation, cells were incubated with either a proteasomal inhibitor, 10 µM MG-132 (Sigma-Aldrich), or a lysosomal inhibitor, 100 µM Chloroquine (Sigma-Aldrich). For the screen of cabozantinib responsive phospho-kinases, the Proteome Profiler Human Phospho-Kinase Array Kit (R&D Systems, Minneapolis, MN, USA) was used with lysates of MDA-MB-231 cells treated with either DMSO or 5 µM cabozantinib for 30 min.
Cell migration and invasion assays
Differences in TN breast cancer cell migration in response to cabozantinib and erlotinib was assessed using wound closure and transwell migration assays. For wound closure assays, MDA-MB-468 or MDA-MB-231 cells were cultured in 12-well dishes to approximately 100% confluency. Under sterile conditions, a vertical wound in the monolayer was created by dragging a 200 µL pipette tip across each well. Cells were then incubated with respective inhibitors (MDA-MB-468 + erlotinib (1–10 µM), MDA-MB-231 + cabozantinib (1–10 µM)] or DMSO control, in combination with or without 10 µg/mL mitomycin C to control for proliferation. Images of 3 low-powered fields of view (LP-FOVs)/well were taken every 12 hours until the wounds in the control wells were fully closed. The mean wound width per FOV was calculated using FIJI ImageJ software. For transwell migration assays, Falcon® transwell inserts (8 µm pore size) were placed in 24-well dishes, coated with gelatin and exposed to culture media (10% FBS) in the bottom well. Cells (5×104 cells/well) were seeded onto the top portion of each transwell chamber and treated with inhibitors, vehicle control (DMSO), and proliferation control (mitomycin C). Following a 24 h incubation at 37 °C, 5% CO2, transwells were fixed with 1% glutaraldehyde and mounted and stained with Vectashield® Antifade Mounting Medium with DAPI (Vector Laboratories). Five high powered (HP)-FOVs were analyzed for each well, and a mean number of migrated cells per FOV was calculated using FIJI ImageJ software. Differences between TN breast cancer cell invasion in response to cabozantinib and erlotinib was assessed using transwells coated with MatrigelTM (15% v/v; BD Science, San Jose, CA, USA), with all other parameters consistent with that described for migration assays.
Statistical analysis
All in vitro experiments were performed with a minimum of three biological replicates, with at least three technical replicates included in each experiment. Quantitative data were compiled from all experiments. Unless otherwise noted, data are presented as the mean ± SD. Statistical analysis was performed using GraphPad Prism 8.0 software (GraphPad Software, San Diego, CA, USA) using analysis of variance (ANOVA) with Tukey post-tests (for comparison between all treated conditions) and non-linear regression (for IC50 dose-curves). Values of P≤0.05 were considered statistically significant.
Results
Expression of EGFR is elevated in TNBC breast cancer patient samples and cell line models, and MDA-MB-468 TNBC cells are responsive to the anti-EGFR inhibitor erlotinib
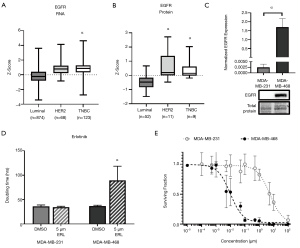
To help define the clinical context, we first evaluated EGFR expression data from TCGA and CPTAC. Analysis of TCGA datasets demonstrated that RNA expression of EGFR is significantly elevated (P<0.05) in TNBC patient samples (z-score: 0.85, n=123) relative to luminal patient samples (z-score: −0.22, n=874) (Figure 1A). Similarly, analysis of CPTAC datasets demonstrated that protein expression of EGFR is also significantly elevated (P<0.05) in TNBC (z-score: 0.40, n=9) relative to luminal breast cancer (z-score: −0.48, n=52) in patient samples (Figure 1B). Moving to experimental studies, we evaluated EGFR expression in our two human TNBC cell line models; MDA-MB-468 and MDA-MB-231. We observed that EGFR was significantly (P<0.05) more expressed in the MDA-MB-468 than the MDA-MB-231 cell line (Figure 1C). We next assessed the response of TNBC cells to the EGFR inhibitor erlotinib with regards to cell growth/proliferation. We observed that the doubling time of MDA-MB-468 cells treated with 5 µM erlotinib (89.3±29.3 h) was significantly increased (P<0.05) relative to DMSO control (36.9±1.7 h) (Figure 1D). However, MDA-MB-231 TNBC cells demonstrated no difference in doubling time in the presence versus absence of inhibitor. To determine the dose-response of TNBC cells to erlotinib, a clonogenic assay was performed. Non-linear regression of the surviving fraction (ratio of number of colonies present: initial seeding density multiplied by the plating efficiency) demonstrated an IC50 =9.705 nM (R2=0.9287) for MDA-MB-468 cells and an IC50 =5.480 µM (R2=0.8205) for MDA-MB-231 cells (Figure 1E). These results suggest that erlotinib is effective at inhibiting growth in the MDA-MB-468 TNBC model (responsive), but not in the MDA-MB-231 model (resistant).
Erlotinib treatment attenuates EGFR expression, phosphorylation and downstream ERK signaling in responsive MDA-MB-468 TNBC cells
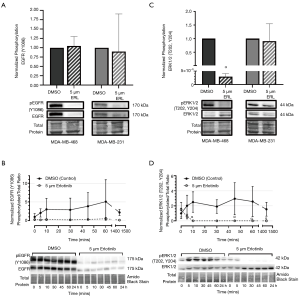
To investigate the mechanistic effects of erlotinib on EGFR signaling, expression and phosphorylation of EGFR and ERK were assessed by immunoblotting of cells treated with erlotinib. Phosphorylated EGFR (Y1086 phospho-site) was significantly reduced (P<0.05) in erlotinib-treated (5 µM) MDA-MB-468 or MDA-MB-231 cells relative to DMSO control (Figure S1A). Unexpectedly, total EGFR expression was also significantly reduced (P<0.05) in both cell line models (Figure S1B), resulting in no change in the ratio of phosphorylated: total protein (Figure 2A). This pattern was also observed in MDA-MB-468 cells over a time course (0–24 hours) of exposure to erlotinib (Figure 2B), with EGFR expression and phosphorylation declining as early as 5 minutes after initial exposure to erlotinib, but no significant difference in the ratio between phosphorylated: total protein across the timepoints. To begin to investigate the potential mechanisms underlying erlotinib-induced attenuation of EGFR expression, we probed degradation pathways mediated by the proteasome and lysosome. Previous studies have suggested that EGFR degradation in cancer cells may be induced by environmental stressors (e.g., chemotherapeutic agents, targeted agents, oxidative stress, etc.) and that this degradation is mediated by canonical recycling pathways (49). To assess the potential contribution of the proteasome, MG-132 (an inhibitor of the 26S proteasome) was incubated with MDA-MB-468 cells treated with either erlotinib or DMSO control. To assess the contribution of the lysosome, the lysosomal inhibitor chloroquine (CQ) was used. We observed that neither MG-132 or CQ had any significant effect on erlotinib-induced attenuation of EGFR expression (Figure S1C,D), indicating that other pathways are likely responsible for EGFR attenuation/degradation. Although the effects of erlotinib on EGFR phosphorylation and expression were unexpected, examination of downstream canonical signaling pathways demonstrated that erlotinib does have has predictable effects on ERK1/2 phosphorylation in responsive MDA-MB-468 TNBC cells. In 5 µM erlotinib-treated MDA-MB-468 cells, phosphorylation levels of ERK1/2 at phospho-sites T202 and Y204 were significantly decreased (P<0.05) relative to DMSO control (Figure 2C). Investigation of the time dependency of erlotinib response revealed that ERK1/2 phosphorylation attenuation (P<0.05) occurs within 10 minutes of exposure to erlotinib and that phosphorylation levels of ERK1/2 remain attenuated, up to 24 h, as long as erlotinib is present (Figure 2D). No significant change in ERK1/2 phosphorylation was observed in non-responsive MDA-MB-231 cells.
Expression of c-Met is elevated in TNBC breast cancer patient samples and cell line models, and MDA-MB-231 TNBC cells are responsive to the anti-c-MET inhibitor cabozantinib
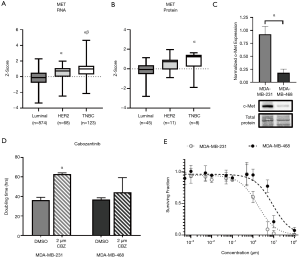
Similar to EGFR, analysis of TCGA datasets demonstrated that RNA expression of c-Met is significantly elevated (P<0.05) in TNBC patient samples (z-score: 0.84, n=123) relative to luminal (z-score: −0.18, n=874) and HER2 (z-score: 0.48, n=68) samples (Figure 3A). Analysis of CPTAC datasets demonstrated that protein expression of c-Met is also significantly elevated (P<0.05) in TNBC (z-score: 0.71, n=8) patient samples relative to luminal samples (z-score: −0.48, n=52) (Figure 3B). Examination of c-Met expression in our two human TNBC cell line models demonstrated elevated expression (P<0.05) in MDA-MB-231 cells relative to MDA-MB-468 cells (Figure 3C). We next assessed the response of TNBC cells to the c-Met inhibitor cabozantinib with regards to cell growth/proliferation. We observed that the doubling time of MDA-MB-231 cells treated with 2 µM cabozantinib (63.0±1.17 h) was significantly increased (P<0.05) relative to control (36.2±2.9 h) (Figure 3D). However, MDA-MB-468 TNBC cells demonstrated no difference in doubling time in the presence versus absence of inhibitor. To determine the dose-response of TNBC cells to cabozantinib, a clonogenic assay was performed. Non-linear regression of the surviving fraction (ratio of number of colonies present: initial seeding density multiplied by the plating efficiency) demonstrated an IC50 =1.682 µM (R2=0.9039) for MDA-MB-231 cells and an IC50 =8.886 µM (R2=0.8464) for MDA-MB-468 cells (Figure 3E). These results suggest that cabozantinib is effective at inhibiting growth in the MDA-MB-231 TNBC model (responsive), but not in the MDA-MB-468 model (resistant).
AKT1 phosphorylation is attenuated in cabozantinib-treated MDA-MB-231 cells
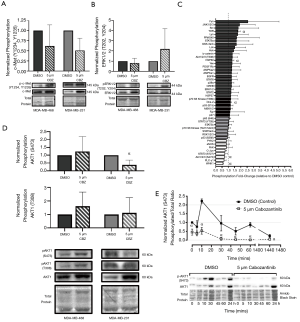
To investigate the mechanistic effects of cabozantinib on c-Met signaling, expression and phosphorylation of c-Met was first assessed by immunoblotting of cells treated with cabozantinib. We did not observe a significant change in phosphorylation of c-Met at Y1230/Y1234/Y1235 (Figure 4A) or canonical ERK1/2 at T202, Y204 (Figure 4B) in either TNBC model. Based on the responsiveness of MDA-MB-231 cells to cabozantinib in the growth/proliferation assays, we wanted to further characterize potential downstream pathways that may be activated. Using a phospho-kinase array, we identified five candidate pathways with significant differences (P<0.05) in phosphorylation between cabozantinib-treated and DMSO-treated controls, including TOR (1.78±0.18), HSP60 (1.33±0.12), Chk-2 (1.20±0.06), AKT1/2/3-S473 (0.72±0.11), and WNK1 (0.59±0.16) (Figure 4C, Figure S2). Given that AKT1 is a canonical downstream effector of Met, we further validated the AKT1 response to cabozantinib using immunoblotting. We observed that phosphorylation of AKT1 at S473 (defined as ratio of phosphorylated: total AKT1 expression) was significantly decreased (P<0.05) in cabozantinib-treated MDA-MB-231 cells relative to DMSO controls (Figure 4D). There were no significant differences in phosphorylation levels of AKT1 T308 between cabozantinib-treated cells and controls (Figure 4D). When investigating the effects over time, we observed that, similar to erlotinib effects in MDA-MB-468 cells, significant decreases (P<0.05) in phosphorylation of Akt1-S473 in cabozantinib-responsive MDA-MB-231 cells occurred after 10 minutes of exposure to inhibitor, and that as long as cabozantinib is present, phosphorylation levels remain significantly (P<0.05) attenuated (Figure 4E).
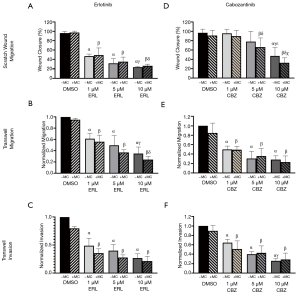
Erlotinib and cabozantinib attenuate migration and invasion of TNBC cells
Finally, given that erlotinib and cabozantinib are used as systemic therapies, we were interested in determining the effects of these inhibitors on biological functions related to metastasis, including migration and invasion. Using scratch wound and transwell migration/invasion assays, we observed that erlotinib (Figure 5A,B,C) significantly inhibited (P<0.05) the migration and invasion of MDA-MB-468 TNBC cells and cabozantinib significantly inhibited (P<0.05) the migration and invasion of MDA-MB-231 TNBC cells (Figure 5D,E,F). Notably, in both cell line models these inhibitory effects were proliferation-independent, suggesting that erlotinib and cabozantinib may be effective at inhibiting multiple functional aspects of disease progression.
Discussion
In this study we explored the potential of EGFR and c-Met as potential targets for treatment of metastatic TNBC, including assessing potential response mechanisms, as there is a lack of effective systemic therapies for TNBC. The anti-EGFR inhibitor erlotinib and anti-c-Met inhibitor cabozantinib may have some clinical activity in controlling disease progression at treatment onset, ultimately, tumors continue to progress and develop resistance to treatment. Thus, studying the underlying mechanisms of c-Met and EGFR inhibition by cabozantinib and erlotinib in TNBC, respectively, may provide understanding to the development of TKI resistance.
We report here that TKIs in select TNBC cell lines have inhibitory effects on cell proliferation, downstream signaling, migration, and invasion. There is evidence supporting that inhibitor effects are mediated by EGFR and c-Met signaling axes, but perhaps not by inactivation of EGFR, due to unexplained loss of expression, nor c-Met, where phosphorylation was not changed in response to TKIs, but through down-regulation of AKT1 activation. We report here potential non-canonical effects of erlotinib and cabozantinib on EGFR and c-Met signaling axes through which these inhibitors inhibit cell proliferation, migration, and invasion.
The increased expression of EGFR and c-Met observed in TNBC patient samples within the TCGA and CPTAC datasets is supported by previous studies, which reported that ~30–50% of TNBC patients overexpressing EGFR or c-Met (22,23,34-36,50,51). The MDA-MB-468 cells are significantly more sensitive to erlotinib than MDA-MB-231 cells as EGFR expression is significantly elevated in the MDA-MB-468 cell line. This is different than what has been reported previously in literature (52) and this is likely due the large difference in EGFR expression between the two cell lines. Additionally, MDA-MB-231 cells have a known KRAS point mutation [COSMIC database (53)] which may also contribute to the observed lack of sensitivity to erlotinib, as KRAS lies within the downstream signaling pathway of EGFR (29,54). In contrast, MDA-MB-231 TNBC cells were sensitive to cabozantinib while MDA-MB-468 were not. This may be due to the increased expression of c-Met in MDA-MB-231 cells compared to MDA-MB-468 cells. Taken together, this highlights the heterogeneity of drug sensitivity and the need to further understand the underlying molecular basis for response versus resistance beyond just RTK expression.
To better understand this heterogeneity, we investigated the effects of the inhibitors on the downstream EGFR and c-Met signaling axis. Based in the anti-proliferative effects of erlotinib in MDA-MB-468 cells, we expected that the ratio of EGFR phosphorylation at Y1086 to be decreased with increasing erlotinib concentration as a reflection of EGFR activation. However, it was difficult to definitively conclude a decrease in EGFR activation in this model due to the corresponding and unexpected loss of EGFR total protein expression induced by erlotinib. This occurred in both cell lines, suggesting that this is not an artifact nor unique to one cell line. This is unusual but not unprecedented, as a previous study by Tsien et al. reported an erlotinib-induced degradation of EGFR in head and neck squamous carcinoma patients (55) also noting an attenuation of activated downstream effectors ERK, AKT, STAT3 and Src in cell lines and patient tumor samples. However, to the best of our knowledge this is the first report of erlotinib-induced EGFR expression loss in TNBC models. Regardless of the mechanism through which erlotinib affects EGFR signaling, the MDA-MB-468 cells remain sensitive to the attenuation of proliferation by erlotinib. ERK1/2 are downstream effectors of EGFR signaling, thus, as might be expected, activation of ERK1/2 via phosphorylation of T202 and Y204 phospho-sites was significantly decreased in response to erlotinib in MDA-MB-468 cells (responsive), but not in MDA-MB-231 cells (resistant). This difference in signaling pathway activation is consistent with the difference in sensitivity observed between the two cell lines. The known activating KRAS mutation in MDA-MB-231 cells could be the cause of the conserved ERK1/2 signaling and lack of sensitivity to erlotinib treatment. Therefore, in responsive MDA-MB-468 cells, erlotinib is likely attenuating ERK1/2 signaling either through inhibition of EGFR activity [the well-studied mechanism of action for erlotinib (56)], loss of EGFR itself as reported here, or a combination of the two. In particular, erlotinib-induced modulation of EGFR expression remains unexplored as a non-canonical mechanism of anti-tumor activity in TNBC. It has been hypothesized that stress-induced cancer cells, including those treated with EGFR-targeting TKIs such as erlotinib and gefitinib, may increase recycling and/or degradation of EGFR (49) via lysosomal or proteasomal mechanisms. Although this was not found to be the case in the current study, other potential mechanisms may be at play including elevated ROS (reactive oxygen species) levels (57), HSP90 inhibition (58,59), HDAC6 (60) and annexin A6 involvement (61). Future studies are aimed at elucidating the underlying reasons for the observed reduction in EGFR expression in response to erlotinib.
We also investigated the downstream effects of cabozantinib on c-Met signaling. We observed a significant attenuation in activation of AKT1, part of the PI3K/AKT signaling pathway, which falls within the c-Met signaling axis (27). Surprisingly, we did not observe significant decreases in phosphorylation of c-Met at Y1234/Y1235 nor in phosphorylation of ERK1/2 at T202/Y204 in cabozantinib-treated MDA-MB-468 and MDA-MB-231 cells, which prompted us to further investigate the underlying reasons for discrepancy between the sensitivity to cabozantinib between the two cell lines. With the phospho-kinase array, we identified downstream effectors that were responsive to cabozantinib treatment in MDA-MB-231 cells and identified several components (TOR, AKT1/2/3) or interactors [HSP60 (62), WNK (63)] of the PI3K/AKT signaling pathway as well as CHK2 which is involved with cell cycle checkpoints and DNA damage repair (64). Given that the involvement of the PI3K/AKT pathway falls within the c-Met signaling axis, we focused on the effects of cabozantinib on AKT1 activation in MDA-MB-231 cells and observed that exposure to cabozantinib to MDA-MB-231 cells led to the attenuation of phosphorylation at S473 phospho-site, but not the T308 phospho-site, which are the two regulatory phospho-sites for AKT1 activation (65). The lack of c-Met inhibition by cabozantinib is not surprising as cabozantinib is known to be a non-specific inhibitor of c-Met and targets several other RTKs involved in cancer signaling (46), through which cabozantinib may be affecting the MDA-MB-231 cells. Therefore, although cabozantinib does not appear to inhibit c-Met activation in MDA-MB-231 cells, it still attenuates activity of a downstream pathway that is involved in cell proliferation, migration and invasion.
In addition to the anti-proliferative effects of the inhibitors, we also observed that MDA-MB-468 cells treated with erlotinib and MDA-MB-231 cells treated with cabozantinib demonstrated reduced cell migration and invasion compared to controls. These findings are supported by previous observations that EGFR and c-Met signaling are involved in cell migration and invasion, where inhibition by erlotinib and cabozantinib attenuate cell migration/invasion (46,48,66,67). Given that these cell lines were originally established from metastatic lesions (68,69), our findings suggest that EGFR and c-Met inhibitors have the potential to have inhibitory effects not only on proliferation, but also on metastatic behavior of TNBC. Future studies using in vivo models are warranted in order to assess if these observed effects on cell migration and invasion translate to metastasis.
In summary, in the current study we demonstrate that erlotinib and cabozantinib have varying anti-proliferative and anti-migratory effects in different TNBC models. Our findings also suggest that the functional effects of cabozantinib and erlotinib occur via inhibition of ERK and PI3K/AKT signaling pathways and suggest that TNBC cells may develop compensatory signaling mechanisms in order to circumvent the anti-cancer effects of these TKIs. Ongoing studies in our laboratory are aimed at investigating these alternative pathways using unbiased phosphoproteomic and kinomic approaches. Elucidation of the underlying mechanisms that define the heterogenous response to TKIs in TNBC could help identify biomarkers to stratify patients for treatment and/or facilitate discovery of targets to attenuate acquired resistance.
Acknowledgments
Funding: This study was funded by a generous donation from the Lloyd Carr-Harris Foundation through the London Health Science Foundation. A.L.A. is supported by the Breast Cancer Society of Canada. C.L. is supported by a Vanier Canada Graduate Scholarship from the Government of Canada.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editor Jacques Raphael for the series “Management of Triple Negative Breast Cancer” published in Precision Cancer Medicine. The article has undergone external peer review.
Reporting Checklist: The authors have completed the Materials Design Analysis Reporting (MDAR) Checklist. Available at: http://dx.doi.org/10.21037/pcm-20-62
Peer Review File: Available at http://dx.doi.org/10.21037/pcm-20-62
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at: http://dx.doi.org/10.21037/pcm-20-62). The series “Management of Triple Negative Breast Cancer” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020;70:7-30. [Crossref] [PubMed]
- Brenner DR, Weir HK, Demers AA, et al. Projected estimates of cancer in Canada in 2020. CMAJ 2020;192:E199-205. [Crossref] [PubMed]
- Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature 2000;406:747-52. [Crossref] [PubMed]
- Sørlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A 2001;98:10869-74. [Crossref] [PubMed]
- Hudis CA. Trastuzumab--Mechanism of Action and Use in Clinical Practice. N Engl J Med 2007;357:39-51. [Crossref] [PubMed]
- Ignatiadis M, Sotiriou C. Luminal breast cancer: From biology to treatment. Nat Rev Clin Oncol 2013;10:494-506. [Crossref] [PubMed]
- Foulkes WD, Smith IE, Reis-Filho JS. Triple-Negative Breast Cancer. N Engl J Med 2010;363:1938-48. [Crossref] [PubMed]
- Zardavas D, Baselga J, Piccart M. Emerging targeted agents in metastatic breast cancer. Nat Rev Clin Oncol 2013;10:191-210. [Crossref] [PubMed]
- Kawalec P, Łopuch S, Mikrut A. Effectiveness of targeted therapy in patients with previously untreated metastatic breast cancer: A systematic review and meta-analysis. Clin Breast Cancer 2015;15:90-100.e1. [Crossref] [PubMed]
- Lang JE, Wecsler JS, Press MF, et al. Molecular markers for breast cancer diagnosis, prognosis and targeted therapy. J Surg Oncol 2015;111:81-90. [Crossref] [PubMed]
- Fallahpour S, Navaneelan T, De P, et al. Breast cancer survival by molecular subtype: a population-based analysis of cancer registry data. CMAJ Open 2017;5:E734-E739. [Crossref] [PubMed]
- Howlader N, Cronin KA, Kurian AW, et al. Differences in Breast Cancer Survival by Molecular Subtypes in the United States. Cancer Epidemiol Biomarkers Prev 2018;27:619-26. [Crossref] [PubMed]
- Druker BJ, Tamura S, Buchdunger E, et al. Effects of a selective inhibitor of the Abl tyrosine kinase on the growth of Bcr-Abl positive cells. Nat Med 1996;2:561-6. [Crossref] [PubMed]
- Lin CR, Chen W, Kruiger W, et al. Expression cloning of human EGF receptor complementary DNA: gene amplification and three related messenger RNA products in A431 cells. Science 1984;224:843-8. [Crossref] [PubMed]
- Coussens L, Yang-Feng T, Liao Y, et al. Tyrosine kinase receptor with extensive homology to EGF receptor shares chromosomal location with neu oncogene. Science 1985;230:1132-9. [Crossref] [PubMed]
- Schechter AL, Hung M, Vaidyanathan L, et al. The neu gene: an erbB-homologous gene distinct from and unlinked to the gene encoding the EGF receptor. Science 1985;229:976-8. [Crossref] [PubMed]
- Blume-Jensen P, Hunter T. Oncogenic kinase signalling. Nature 2001;411:355-65. [Crossref] [PubMed]
- Lemmon MA, Schlessinger J. Cell Signaling by Receptor Tyrosine Kinases. Cell 2010;141:1117-34. [Crossref] [PubMed]
- Weinstein IB, Joe A, Felsher D. Oncogene Addiction. Cancer Res 2008;68:3077-80. [Crossref] [PubMed]
- Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or Carboplatin-Paclitaxel in Pulmonary Adenocarcinoma. N Engl J Med 2009;361:947-57. [Crossref] [PubMed]
- Cappuzzo F, Ciuleanu T, Stelmakh L, et al. Erlotinib as maintenance treatment in advanced non-small-cell lung cancer: a multicentre, randomised, placebo-controlled phase 3 study. Lancet Oncol 2010;11:521-9. [Crossref] [PubMed]
- Corkery B, Crown J, Clynes M, et al. Epidermal growth factor receptor as a potential therapeutic target in triple-negative breast cancer. Ann Oncol 2009;20:862-7. [Crossref] [PubMed]
- Lehmann BD, Bauer JJ, Chen X, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest 2011;121:2750-67. [Crossref] [PubMed]
- Gelmon K, Dent R, Mackey JR, et al. Targeting triple-negative breast cancer: Optimising therapeutic outcomes. Ann Oncol 2012;23:2223-34. [Crossref] [PubMed]
- Engelman JA, Zejnullahu K, Mitsudomi T, et al. MET Amplification Leads to Gefitinib Resistance in Lung Cancer by Activating ERBB3 Signaling. Science 2007;316:1039-43. [Crossref] [PubMed]
- Lai AZ, Abella JV, Park M. Crosstalk in Met receptor oncogenesis. Trends Cell Biol 2009;19:542-51. [Crossref] [PubMed]
- Gentile A, Trusolino L, Comoglio PM. The Met tyrosine kinase receptor in development and cancer. Cancer Metastasis Rev 2008;27:85-94. [Crossref] [PubMed]
- Boccaccio C, Andò M, Tamagnone L, et al. Induction of epithelial tubules by growth factor HGF depends on the STAT pathway. Nature 1998;391:285-8. [Crossref] [PubMed]
- Marshall CJ. Specificity of receptor tyrosine kinase signaling: Transient versus sustained extracellular signal-regulated kinase activation. Cell 1995;80:179-85. [Crossref] [PubMed]
- Salomon DS, Brandta R, Ciardiellob F, et al. Epidermal growth factor-related peptides and their receptors in human malignancies. Crit Rev Oncol Hematol 1995;19:183-232. [Crossref] [PubMed]
- Lurje G, Lenz HJ. EGFR signaling and drug discovery. Oncology 2009;77:400-10. [Crossref] [PubMed]
- Martinazzi M, Crivelli F, Zampatti C, et al. Epidermal growth factor receptor immunohistochemistry in different histological types of infiltrating breast carcinoma. J Clin Pathol 1993;46:1009-10. [Crossref] [PubMed]
- Sainsbury JR, Farndon JR, Needham GK, et al. Epidermal-growth-factor receptor status as predictor of early recurrence of and death from breast cancer. Lancet 1987;1:1398-402. [PubMed]
- Dent R, Trudeau M, Pritchard KI, et al. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res 2007;13:4429-34. [Crossref] [PubMed]
- Burness ML, Grushko TA, Olopade OI. Epidermal Growth Factor Receptor in Triple-Negative and Basal-Like Breast Cancer. Cancer J 2010;16:23-32. [Crossref] [PubMed]
- Guérin M, Gabillot M, Mathieu MC, et al. Structure and expression of c-erbB-2 and EGF receptor genes in inflammatory and non-inflammatory breast cancer: prognostic significance. Int J Cancer 1989;43:201-8. [Crossref] [PubMed]
- Sameni M, Tovar EA, Essenburg CJ, et al. Cabozantinib (XL184) Inhibits Growth and Invasion of Preclinical TNBC Models. Clin Cancer Res 2016;22:923-34. [Crossref] [PubMed]
- Schöffski P, Gordon M, Smith DC, et al. Phase II randomised discontinuation trial of cabozantinib in patients with advanced solid tumours. Eur J Cancer 2017;86:296-304. [Crossref] [PubMed]
- Tolaney SM, Nechushtan H, Ron IG, et al. Cabozantinib for metastatic breast carcinoma: results of a phase II placebo-controlled randomized discontinuation study. Breast Cancer Res Treat 2016;160:305-12. [Crossref] [PubMed]
- Tan AR, Yang X, Hewitt SM, et al. Evaluation of biologic end points and pharmacokinetics in patients with metastatic breast cancer after treatment with erlotinib, an epidermal growth factor receptor tyrosine kinase inhibitor. J Clin Oncol 2004;22:3080-90. [Crossref] [PubMed]
- Dickler MN, Rugo HS, Eberle CA, et al. A Phase II Trial of Erlotinib in Combination with Bevacizumab in Patients with Metastatic Breast Cancer. Clin Cancer Res 2008;14:7878-83. [Crossref] [PubMed]
- Ueno NT, Zhang D. Targeting EGFR in Triple Negative Breast Cancer. J Cancer 2011;2:324-8. [Crossref] [PubMed]
- Sharma P, Khan Q, Kimler B, et al. Abstract P1-11-07: Results of a Phase II Study of Neoadjuvant Platinum/Taxane Based Chemotherapy and Erlotinib for Triple Negative Breast Cancer. Cancer Res 2010;70:P1-11-07.
- Maurer MA, Kalinsky K, Crew K, et al. Abstract 28: Phase I trial of combined temsirolimus, erlotinib, and cisplatin in advanced solid tumors. Cancer Res 2013;73:28.
- Xiang Q, Chen W, Ren M, et al. Cabozantinib suppresses tumor growth and metastasis in hepatocellular carcinoma by a dual blockade of VEGFR2 and MET. Clin Cancer Res 2014;20:2959-70. [Crossref] [PubMed]
- Yakes FM, Chen J, Tan J, et al. Cabozantinib (XL184), a Novel MET and VEGFR2 Inhibitor, Simultaneously Suppresses Metastasis, Angiogenesis, and Tumor Growth. Mol Cancer Ther 2011;10:2298-308. [Crossref] [PubMed]
- Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017;19:649-58. [Crossref] [PubMed]
- Wang XY, Smith DI, Frederick L, et al. Analysis of EGF receptor amplicons reveals amplification of multiple expressed sequences. Oncogene 1998;16:191-5. [Crossref] [PubMed]
- Tan X, Lambert PF, Rapraeger AC, et al. Stress-Induced EGFR Trafficking: Mechanisms, Functions, and Therapeutic Implications. Trends Cell Biol 2016;26:352-66. [Crossref] [PubMed]
- Graveel CR, DeGroot JD, Su Y, et al. Met induces diverse mammary carcinomas in mice and is associated with human basal breast cancer. Proc Natl Acad Sci 2009;106:12909-14. [Crossref] [PubMed]
- Ponzo MG, Lesurf R, Petkiewicz S, et al. Met induces mammary tumors with diverse histologies and is associated with poor outcome and human basal breast cancer. Proc Natl Acad Sci 2009;106:12903-8. [Crossref] [PubMed]
- El Guerrab A, Bamdad M, Kwiatkowski F, et al. Anti-EGFR monoclonal antibodies and EGFR tyrosine kinase inhibitors as combination therapy for triple-negative breast cancer. Oncotarget 2016;7:73618-37. [Crossref] [PubMed]
- Bamford S, Dawson E, Forbes S, et al. The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. Br J Cancer 2004;91:355-8. [Crossref] [PubMed]
- Downward J. Targeting RAS signalling pathways in cancer therapy Nat Rev Cancer 2003;3:11-22. [Crossref] [PubMed]
- Tsien CI, Nyati MK, Ahsan A, et al. Effect of erlotinib on epidermal growth factor receptor and downstream signaling in oral cavity squamous cell carcinoma. Head Neck 2013;35:1323-30. [Crossref] [PubMed]
- Raymond E, Faivre S, Armand JP. Epidermal growth factor receptor tyrosine kinase as a target for anticancer therapy. Drugs 2000;60:15-23; discussion 41-2. [Crossref] [PubMed]
- Leung ELH, Fan XX, Wong MP, et al. Targeting tyrosine kinase inhibitor-resistant non-small cell lung cancer by inducing epidermal growth factor receptor degradation via methionine 790 oxidation. Antioxid Redox Signal 2016;24:263-79. [Crossref] [PubMed]
- Hong YS, Jang WJ, Chun KS, et al. Hsp90 inhibition by WK88-1 potently suppresses the growth of gefitinib-resistant H1975 cells harboring the T790M mutation in EGFR. Oncol Rep 2014;31:2619-24. [Crossref] [PubMed]
- Sawai A, Chandarlapaty S, Greulich H, et al. Inhibition of Hsp90 down-regulates mutant epidermal growth factor receptor (EGFR) expression and sensitizes EGFR mutant tumors to paclitaxel. Cancer Res 2008;68:589-96. [Crossref] [PubMed]
- Gao YS, Hubbert CC, Yao TP. The microtubule-associated histone deacetylase 6 (HDAC6) regulates epidermal growth factor receptor (EGFR) endocytic trafficking and degradation. J Biol Chem 2010;285:11219-26. [Crossref] [PubMed]
- Koumangoye RB, Nangami GN, Thompson PD, et al. Reduced annexin A6 expression promotes the degradation of activated epidermal growth factor receptor and sensitizes invasive breast cancer cells to EGFR-targeted tyrosine kinase inhibitors. Mol Cancer 2013;12:167. [Crossref] [PubMed]
- Yan FQ, Wang JQ, Tsai YP, et al. HSP60 overexpression increases the protein levels of the p110α subunit of phosphoinositide 3-kinase and c-Myc. Clin Exp Pharmacol Physiol 2015;42:1092-7. [Crossref] [PubMed]
- Gallolu Kankanamalage S, Karra AS, Cobb MH. WNK pathways in cancer signaling networks. Cell Commun Signal 2018;16:72. [Crossref] [PubMed]
- Yang S, Kuo C, Bisi JE, et al. PML-dependent apoptosis after DNA damage is regulated by the checkpoint kinase hCds1/Chk2. Nat Cell Biol 2002;4:865-70. [Crossref] [PubMed]
- Alessi DR, Andjelkovic M, Caudwell B, et al. Mechanism of activation of protein kinase B by insulin and IGF-1. EMBO J 1996;15:6541-51. [Crossref] [PubMed]
- Eccles SA. The epidermal growth factor receptor/Erb-B/HER family in normal and malignant breast biology. Int J Dev Biol 2011;55:685-96. [Crossref] [PubMed]
- Birchmeier C, Birchmeier W, Gherardi E, et al. Met, metastasis, motility and more. Nat Rev Mol Cell Biol 2003;4:915-25. [Crossref] [PubMed]
- Cailleau R, Olivé M, Cruciger QVJ. Long-term human breast carcinoma cell lines of metastatic origin: Preliminary characterization. In Vitro 1978;14:911-5. [Crossref] [PubMed]
- Engel LW, Young NA. Human Breast Carcinoma Cells in Continuous Culture: A Review. Cancer Res 1978;38:4327-39. [PubMed]
Cite this article as: Lefebvre C, Allan AL. Anti-proliferative and anti-migratory effects of EGFR and c-Met tyrosine kinase inhibitors in triple negative breast cancer cells. Precis Cancer Med 2021;4:2.

