
Digging deep or spreading wide?—a comparison of polymerase chain reaction and sequencing approaches in the analysis of circulating tumor DNA
Introduction
Liquid biopsy, the non-invasive analysis of circulating tumor DNA (ctDNA) in plasma, is quickly becoming a versatile and promising method in clinical oncology (1). Practical applications for lung cancer are currently confined to non-invasive analysis of the tumor genome, with the goal of establishing or adjusting therapy (2). For instance, when a surgical biopsy is impossible or unsuccessful, EGFR mutations can be detected in plasma, prompting for treatment with specific tyrosine kinase inhibitors (3-5). The same test can then be used to monitor treatment efficiency and detect the appearance of resistance-causing mutations (6). An example of the latter is the well-known p.Thr790Met (aka T790M), which alters the conformation of the ATPase site in EGFR and prevents binding of first- or second-generation tyrosine kinase inhibitors (7). The early detection of p.Thr790Met is key to a timely switch to a family of third-generation EGFR inhibitors specific for the presence of this mutation (8). Such a therapeutic strategy is greatly facilitated by liquid biopsy, since regular blood draws are much less of a burden for the patient than lung biopsies, and detection of p.Thr790Met in ctDNA was shown to occur several months before tumor growth becomes detectable by imaging (9). Other applications of ctDNA analysis, most of them still in development, include non-invasive appraisal and follow-up of tumor mutation burden, verification of the efficiency of surgery or radiotherapy, residual disease monitoring and early detection of relapses (3,10).
For all its promises, ctDNA analysis is technically highly challenging, owing to the low amounts of circulating cell-free DNA in plasma [typically in the order of 10 ng DNA per mL plasma (1) and unpublished observations] and to the fact that ctDNA often accounts for only a minute fraction of cell-free DNA (11), keeping in mind that the clinically relevant threshold might be as low as 0.1%. Since 10 ng represent roughly 3,000 copies of the human genome, detection of a mutation at 0.1% frequency implies to reliably detect 3 mutant molecules per ml plasma. This technical feat necessitates highly optimized laboratory procedures, from plasma collection to DNA extraction, to mutation detection. While it is relatively easy to maximize the amount of plasma by drawing more blood (within reasonable limits) and to optimize DNA yield with dedicated extraction methods, detection of mutations at very low frequencies remains highly challenging (1,12).
Traditionally, mutation detection methods follow two distinct strategies: going deep or spreading wide. The former is used when it is important to detect even minute amounts of a precise mutation, e.g., a resistance-causing mutation, or a mutation that was detected in the patient prior to therapy and serves as a highly sensitive and specific marker for relapses (10,13). In such cases, only one or a few genomic positions are interrogated using highly sensitive techniques that can reliably detect only a few molecules of mutant DNA. On the other hand, there are situations in which the mere presence of mutations, rather than their precise identities, is clinically relevant and the search can be extended to wider genomic regions: an entire gene, a panel of genes, the entire human exome or even the whole genome. Such methods allow the detection of practically any mutation in the target region(s), without prior knowledge of its existence. In addition, since most tumors contain multiple mutations, a wider target area increases the probability of detecting at least one mutation, thereby improving total sensitivity even if ‘per mutation’ sensitivity is lower with sequencing than with polymerase chain reaction (PCR) (14). Furthermore, this approach may allow the discovery of novel resistance mechanisms and is the only available when one is interested in the number of mutations rather than in their precise identity, for instance when estimating tumor mutation burden to evaluate the chances of success of immunotherapy (15,16).
PCR is potentially the most sensitive mutation detection technique: theoretically, given enough PCR cycles, even a single DNA molecule can be amplified to the microgram range, so it can easily be analyzed by standard laboratory procedures. PCR has limitations, though, and among these is the difficulty to quantify the number of original DNA molecules in the reaction: after a given number of cycles the DNA polymerase becomes the limiting factor in the reaction which tends towards a plateau. To obtain precise quantification of the initial material, one must either stop the PCR reaction in its exponential phase, which is difficult to predict, or preferably follow it in real time so one can analyze the exponential phase no matter when it occurs (17).
Real-time PCR, which monitors DNA accumulation with fluorescent dyes, is a well-established method to quantify the abundance of a mutation in a DNA sample. It implies a way to distinguish mutant DNA from wild-type, which can be achieved either by amplification with primers specific for the presence or absence of a mutation (as in ARMS-PCR (18)) or by detection with probes specific for mutant or wild-type amplicons [e.g., Taqman assay (19)].
Both approaches suffer from the same limitation in specificity: particularly with a single-nucleotide mutation, it is impossible to design 100% specific primers or probes. There is always a small amount of cross-binding to the wild-type sequence, which translates into low levels of signal even in the absence of mutation. The conundrum then becomes to decide whether a low signal in late PCR cycles is due to non-specific binding or to the presence of a minute amount of mutant molecules (19). In other words, when mutations frequencies become too low, they cannot be distinguished from background PCR noise.
Digital PCR was designed to overcome this problem. In this approach, thousands of PCR reactions are performed in parallel, either in an emulsion of microdroplets or in microwells engraved on (for instance) a silicon microchip (20). The original sample is diluted enough that there is never more than one DNA molecule per well/droplet. A large number of PCR cycles follows but instead of monitoring product accumulation with time, one assesses the final result, generally with a pair of differently colored probes specific for mutant and wild-type sequences. The percentage of mutation in the original sample is inferred from the number of wells/droplets that light up with the mutant probe, versus the wild-type (20). Because there was only a single DNA molecule to start with, a minor amount of non-specific signal in a well/droplet can be safely ignored.
The main drawback of digital PCR is that, since all wells/droplets must interrogate the same mutation, it is limited to one mutation per assay. Techniques have been proposed to multiplex several amplicons in the same assay, by using probes of different colors or different intensities, but these are limited to a few mutations per assay (21). By contrast, real-time PCR can interrogate a different mutation in each well, so potentially 96 or even 384 mutations in parallel, depending on the capacity of the thermocycler.
In this paper, we wish to review the main techniques currently available for mutation detection in ctDNA, and illustrate our purpose with comparative experiments performed in our laboratory. Generally speaking, the “narrow range, high depth” techniques involve PCR, whether digital PCR or real-time PCR, while “wide-range, shallow depth” techniques imply high throughput sequencing. However, a recent technical improvement, molecular barcodes, is now bringing the sensitivity of sequencing close to that of PCR and might become the method of choice for ctDNA analysis (22).
As test material for both techniques, we employed reference DNA samples containing a cocktail of 8 mutations in well-defined proportions (Horizon Discoveries Ltd., see Methods). The main drawback of this material is that it is not genuine circulating cell-free DNA, but rather genomic DNA fragmented to a length approximating that of cell-free DNA. We thus repeated our validation tests with cell-free DNA from patients carrying at least two known mutations, diluted with cell-free DNA from a healthy carrier to reduce mutation frequencies to more challenging levels.
Methods
Samples
DNA was extracted from anonymized plasma samples from patients with lung or colon cancer and from a healthy donor, as previously described (23). No patient-related information was transmitted to our laboratory, other than the location of the tumor (lung or colon) and the mutations found in it during routine diagnostic screening by high-throughput sequencing in another laboratory. Patient cell-free DNA was serially diluted in cell-free DNA from the healthy donor so as to lower the mutation frequency: 1/10, 1/40, 1/160 and 1/640 for ARMS-PCR and 1/3, 1/9, 1/27, 1/81, 1/243 for sequencing. Upon sequencing, the dilution factor was verified with single nucleotide polymorphisms that differed in the patient and the healthy donor.
Reference DNA was bought from Horizon Discoveries Ltd and used to validate both PCR and sequencing. It consists in 1 wild-type control and 3 samples containing 8 mutations in well-defined concentrations: EGFR:L858R, EGFR:T790M, EGFR:E746_A750del and EGFR:V769_D770insASV at 5%, 1% and 0.1%, as well as KRAS:G12D, NRAS:Q61K, NRAS:A59T and PIK3CA:E545K at 6.3%, 1.3% and 0.13%. Three of these mutations are detectable with the theraScreen kit (EGFR:L858R, EGFR:T790M and EGFR:E746_A750del), and all of them by sequencing. The electrophoretic profile of this material (not shown) reveals that it is not genuine cell-free DNA, although it is comparable in size.
ARMS-PCR
Mutations were detected by real-time PCR with the theraScreen EGFR kit (Qiagen), based on the “amplification-refractory mutation system” (ARMS) principle (18). The kit consists of 4 parallel PCR reactions: control amplicon, T790M, L858R and a cocktail of 19 EGFR exon 19 indels. A standard curve was built by serial dilution of the positive control included in the kit. PCR was monitored with a LC480 thermocyclers (Roche), signal was quantified with both the “fit points threshold” and the “2nd derivative maximum” methods. Mutation percentages were calculated from the difference between a given amplicon and the control amplicon, deriving PCR efficiency from the standard curve.
Sequencing
Libraries were prepared using the QiaSeq Lung cancer panel (Qiagen), which includes 4,149 primers targeting the exons of 72 cancer genes, with a total sequencing footprint of about 500 kb. Molecular barcodes are introduced as part of an adapter ligated to each DNA fragment prior to primer extension. Libraries were sequenced as 2×150 nt on a NextSeq 500 sequencer (Illumina), aiming for a sequencing depth of 3,000–5,000× for most samples and 10,000–15,000× for extreme dilutions. Molecular depth after barcode deduplication and filtering was 550–750× for plasma samples (10 ng cell-free DNA input) and 800–1,200× for reference material (40 ng DNA input). Data was analyzed with smCounter v.2 (24) on a local Unix cluster.
Results
PCR approaches
In our laboratory, we elected to validate a real-time PCR technique, namely the EGFR theraScreen test, which allows for the simultaneous detection of 21 EGFR mutations including p.Leu858Arg which is responsible for approximately 40% of EGFR-related cases (25). Another 40% of cases carry insertion/deletions in EGFR exon 19 and the theraScreen kit comprises a cocktail of primers targeting 19 well-known indels (thus the kit can detect that one of these is present, but cannot determine which). Finally, it detects the p.Thr790Met mutation, a well-known cause of resistance to tyrosine kinase inhibitor therapy (7).
The reference material from Horizon Discoveries Ltd consists of 3 DNA samples, each containing p.Leu858Arg, p.Thr790Met and an exon 19 indel at a given percentage: 5%, 1% and 0.1%. All 3 mutations were detected by real-time PCR at the 3 concentrations with an excellent linearity: R2 >0.98 (Figure 1A), whereas no signal was observed in the wild-type control included with the reference material. However, it should be noted that mutation frequencies were underestimated, particularly for the p.Leu858Arg mutation. This may be due to amplicon-dependent variations in PCR efficiency: if the amplicon containing p.Leu858Arg amplified less well than the control amplicon, its concentration, determined by comparison with the control amplicon, would be systematically underestimated. This is a drawback of any real-time PCR system but, since the resulting bias is systematic, it can be taken into account and does not prevent comparisons between samples (e.g., for patient follow-up).
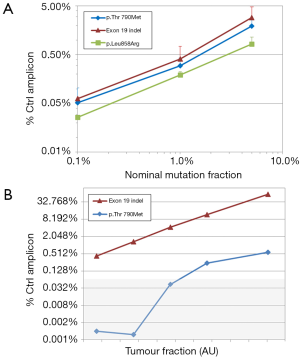
We then analyzed DNA extracted from the plasma of a patient whose lung tumor contained two EGFR mutations: an indel in exon 19, likely the driver mutation, and the resistance-inducing mutation p.Thr790Met at a lower frequency, in line with the hypothesis that p.Thr790Met appeared in a subclone of the main tumor. The corresponding frequencies appraised by real-time PCR were 13.7% and 1.3% respectively. Cell-free DNA from this patient was then serially diluted down to 1/640 with cell-free DNA from a healthy donor, theoretically bringing the frequency of the two mutations down to 0.02% and 0.002%, respectively. Real-time PCR was able to detect the exon 19 indel at all dilutions, in a highly linear manner: R2 >0.99 (Figure 1B). The less abundant p.Thr790Met was detected down to a dilution of 1/40, which theoretically corresponds to a 0.03% mutation frequency. Unsurprisingly, no signal was detected at dilutions 1/160 and 1/640, respectively corresponding to mutation frequencies of 0.008% and 0.002%: Given the initial amount of DNA in the reaction (10 ng) these frequencies theoretically result in approximately 0.2 and 0.06 mutant DNA molecules per reaction.
Sequencing approaches
We saw that, while digital PCR interrogates only one genomic position, real time PCR can address several dozen; high throughput sequencing pushes this limit even further, since it has the potential to interrogate thousands, or even millions of genomic positions. In theory, the upper limit is the entire genome, however, because a high sequencing depth (typically several thousand reads) is required to detect low frequency mutations, sequencing costs become unaffordable for routine analysis if the target area is too large. In practice, this requires targeted sequencing, in which only a selection of genes or genomic regions are isolated and sequenced. The isolation procedure can involve either multiplexed specific PCR reactions or hybridization and capture with a pool of specific probes (1). The panel used for this study having a 500 kb footprint, we aimed for an average sequencing depth of 5,000×, implying about 7 million reads per sample, except for samples with expected low mutations frequencies for which we increased sequencing depth up to 14,000×.
The main problem with sequencing is the high background caused by PCR errors and sequencing mistakes, which makes it difficult to identify low-frequency mutations. A greater sequencing depth increases the probability of detecting a low-frequency mutation, but does not reduce the signal-to-noise ratio. However, a recent technological improvement has brought sensitivity of sequencing into a range comparable with that of real-time PCR. It involves ligating a unique DNA sequence to individual DNA fragments prior to any amplification. This so-called molecular barcode (aka molecular tag or UMI: unique molecular identifier) is sequenced together with the fragment it is attached to and allows the software to determine from which original molecule each sequence read came from (22). This information can be leveraged during analysis, for instance by aggregating all sequence reads corresponding to the same DNA molecule into a consensus sequence, in which each position is assigned the nucleotide most frequently observed within the read group (26). With this strategy, or similar ones, the background error rate can be reduced significantly and allows for the detection of mutations at frequencies inferior to 1%, sometimes even less than 0.1%.
Among the 8 mutations present in the reference material from Horizon Discoveries Ltd., 6 were detected by sequencing in a highly linear manner, down to the lowest frequency, 0.1% (Figure 2A). The mutation frequencies appraised by counting distinct molecular barcodes, i.e., distinct initial DNA molecules, were in excellent agreement with values supplied by the manufacturer: 5%, 1% and 0.1% for EGFR mutations, 6.3%, 1.3% and 0.13% for the other mutations. The notable exception were the two indels in EGFR, which were not detectable in the 0.1% standard, while their frequency in the 5% standard was markedly underestimated (1.75% and 2.42%). We were able to track this poor performance down to a bioinformatic alignment problem: when an indel happens to be located near the end of a DNA molecule, alignment stops at the indel and does not resume afterwards, as there is not enough sequence data left to unambiguously map the end of the read to a genomic position. In this instance, sequencing proved weaker than PCR, although one could probably address this particular problem by improving the alignment software.
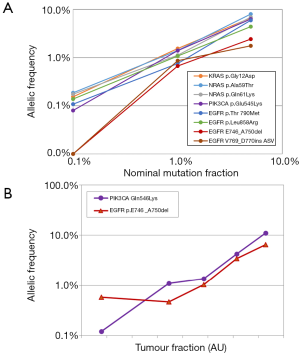
We then extended our validation to genuine ctDNA, from a colon cancer patient whose plasma contained two mutations, PIK3CA:p.Gln546Lys and EGFR:p.Glu746_Ala750del (Figure 2B). In the original sample, the respective frequencies of these mutations, appraised by counting barcodes, were 16.3% and 7.9%. This sample was then serially diluted with cell-free DNA from a healthy donor for final theoretical mutation frequencies of 0.07% and 0.03% respectively. Both mutations were detected at all dilutions with good linearity: R2 >0.95. The exception was the EGFR indel for which the 1/243 revealed the same apparent frequency as the 1/81 dilution. The unexpectedly high value of the last data point might be attributed to stochastic variation: at such a dilution and given the DNA input, one would expect only 3 mutant molecules in this sample.
It should be noted that the above detection was performed with a “white-list” strategy, i.e., looking for sequence alterations corresponding to a list of predefined mutations. This can be viewed as a software equivalent of PCR approaches, in that only predefined mutations can be detected, although in this case the number of predefined mutations is not limited. To detect all mutations, including novel ones, the software must distinguish genuine mutations from background noise, a much more difficult task. In our sequencing experiments, the software provided by the supplier of the molecular barcode library system, smCounter (24), successfully called all mutations down to a frequency of 1%. However at a 0.1% mutation frequency only one mutation was called, NRAS:p.Gln61Lys. In dilutions of patient’s DNA, smCounter detected the PIK3CA mutation down to a frequency of 1.1% and the EGFR deletion down to 1.0%.
Discussion
With the purpose of validating these assays for routine clinical applications, we compared a real-time PCR assay and a high-throughput sequencing method for their ability to detect mutations in ctDNA. To this end, we first used reference control material, in which 8 well-defined mutations are included at precise mutation frequencies. As this material is not genuine circulating cell-free DNA, we also performed serial dilutions of cell-free DNA from a patient into cell-free DNA from a healthy donor.
The real-time PCR assay displayed 100% specificity (no false positive calls, neither in reference material, nor in cell-free DNA dilutions). In reference material, sensitivity was 100% down to 0.1% mutation frequency. With actual cell-free DNA, the detection limit was 0.04%.
High throughput sequencing was almost, but not quite as performant: in reference material it detected all mutations down to 1% mutation frequency, but only 6 out of 8 at 0.1% frequency. The missing two mutations were indels and their non-detection was traced down to an alignment problem. With cell-free DNA, the two mutations sought were detected down to a frequency inferior to 0.1%, although frequency estimation became imprecise at these low values. Detection was achieved using a while-list approach, which focuses on a list of predefined mutations or hotspots. When dispensing with such a list, the variant calling software only detected all mutations down to a frequency of 1%. Specificity was 100%, as no false positive calls were issued by the software.
Our results show that both methods are valid alternatives for ctDNA analysis although, as expected, real-time PCR was slightly more sensitive than sequencing, even with molecular barcodes. Unsurprisingly, other studies came to the same conclusion when applying these tests to diagnostic cohorts, and reported a small number of cases where a mutation was detected by PCR that had not been identified by sequencing (27-29). Interestingly, at least one major center questioned the validity of such results, as 3 out of 4 were not in concordance with the patients’ clinical history, or with the findings in the primary tumor (e.g., detection of a T790M resistance mutation in the absence of EGFR activating mutations) (27).
In theory, digital PCR is even more sensitive than real-time PCR, a fact that was experimentally verified by others (30-32). However, this gain in sensitivity is probably not relevant in the case of liquid biopsy, as the limiting factor in not sensitivity but rather the number of mutant molecules in the original sample. For instance, Zhang et al. demonstrated that digital PCR was able to detect the EGFR:T790M mutation down to 6 copies per assay, while this was not the case with real-time PCR (31). However, with a threshold of 2 events per assay, the probability that a sample expected to contain 6 copies actually contains less than the threshold, is around 6% (Poisson expression). This implies that even a “perfect” assay cannot achieve 100% sensitivity at such low mutation frequencies.
Perhaps for this reason, Watanabe et al. decided on a detection threshold of 10 events per assay, which implied using 50 ng DNA per assay to reach a sensitivity of 0.032% (33). While this was feasible in their case, a study of tumor samples, it would me much more difficult to achieve with ctDNA. In clinical practice, the total amount of cell-free DNA that can be recovered from a single blood draw varies considerably (9), but is often inferior to 30 ng, sometimes as low as 5–10 ng. Based on our experience in the past 3 years (unpublished), a minimum collection of 40 mL blood would be required to guarantee a yield of 50 ng DNA in every case. Thus, there is no point in using a method that can detect one mutant molecule in a million if one can never start from more than 10,000 molecules.
This may be one of the reasons why, to our knowledge, the only commercially available ctDNA tests with either FDA approval or CE-IVD certification are based on real-time PCR. Since one cannot realistically expect to start with more than 10,000 DNA molecules in routine testing, digital PCR might be viewed as an intellectually appealing but unnecessary luxury that bears the extra limitation of testing only one mutation at a time. The latter limitation may soon be overcome, though. Techniques have been described to multiplex several amplicons in the same digital PCR assay, either by using dyes of different intensities (21), by labelling probes with various combinations of dyes (34), or by using pools of primers addressing similar mutations (e.g., 4 nucleotide insertions) at the same genomic location (35). In addition, recent engineering developments promise novel digital PCR machines with the ability to perform 24 parallel reactions of 26,000 microwells, or 96 reactions of 8,500 microwells in dedicated microfluidics microtiter plate (Qiagen, personal communication).
A critical factor, however, will remain the ability to recover and analyze the few mutant molecules that are present in a blood sample when mutation frequency is as low as 0.1%. In this respect, we noticed that sequencing methods are far from optimal: when comparing the total number of barcodes sequenced with the theoretical number of DNA molecules in the original DNA sample, we realized that only about 25% of the original molecules had been sequenced. Because the variant calling software we used arbitrarily requires at least 3 mutant molecules to call a mutation (24), the theoretical limit of detection for this sequencing method becomes 12 molecules per sample. In addition, this suboptimal yield resulted in molecular depths of 1,000× or less in most cases, thereby implying a granularity of at best 0.1%. Molecular yield could be brought up to around 50% by using a hybridization-based system, but this did not improve detection sensitivity (data not shown). When using real-time PCR, our serial dilution experiments reliably detected a mutation when 36 mutant molecules were present in the reaction, but signal was lost at 9 molecules, implying that, here also, molecular recovery is incomplete. In this respect, there is room for improvement with both approaches and, hopefully, technological progress can optimize recovery in the near future. While it will never be possible to overcome the problem of stochastic variations leading to the complete absence of target molecules in a sample, we should at least thrive to detect 100% of those molecules that are present.
Conclusions
We conclude that both real-time PCR and high throughput sequencing with molecular barcodes are appropriate methods for the analysis of ctDNA, notably in the context of lung or colon cancer. In our hands, real-time PCR proved slightly more sensitive and thus more suited to answer a precise question (such as the emergence of EGFR:p.ThrT790Met). By contrast, sequencing covers a wider area, in our case allowing for mutation detection in 72 genes, and would thus be the method of choice in most situations. It is worth mentioning that PCR is much faster than sequencing (a few hours versus a couple of days) and substantially cheaper. Therefore, it is important for a diagnostic laboratory to validate both approaches, so that one can select the one or the other depending on the clinical situation and the question being asked.
Acknowledgments
The author wishes to thank all members of the GTATC (Groupe de Travail sur l’ADN Tumoral Circulant) for constant support and stimulating discussion. I am grateful to Dr. Thibaud Koessler, Dr. Alfredo Addeo and Dr. Tanguy Araud for providing samples, and particularly to the anonymous patients who donated blood.
Funding: The study was funded out of the laboratory method development fund.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Alfredo Addeo and Giuseppe Banna) for the Series “Non-Small Cell Lung Cancer” published in Precision Cancer Medicine. The article was sent for external peer review organized by the Guest Editors and the editorial office.
Conflicts of Interest: The author has completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/pcm.2020.03.01). The Series “Non-Small Cell Lung Cancer” was commissioned by the editorial office without any funding or sponsorship. The author has no conflicts of interest to declare.
Ethical Statement: The author is accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). No ethical approval required according to local laws, as mutations sought in plasma were already known to exist in the tumor. The patient and the healthy donor consented to donate blood for the purpose of method comparison and development. Patients’ samples were anonymized and transmitted to the laboratory without any information pertaining to the patient, other than location of the primary tumor (lung or colon) and the mutations it contained.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Koessler T, Addeo A, Nouspikel T. Implementing circulating tumor DNA analysis in a clinical laboratory: A user manual. Adv Clin Chem 2019;89:131-88. [Crossref] [PubMed]
- Bernabé R, Hickson N, Wallace A, et al. What do we need to make circulating tumour DNA (ctDNA) a routine diagnostic test in lung cancer? Eur J Cancer 2017;81:66-73. [Crossref] [PubMed]
- Diaz LA, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol 2014;32:579-86. [Crossref] [PubMed]
- Zhou C, Wu YL, Chen G, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol 2011;12:735-42. [Crossref] [PubMed]
- Reck M, Hagiwara K, Han B, et al. ctDNA Determination of EGFR Mutation Status in European and Japanese Patients with Advanced NSCLC: The ASSESS Study. J Thorac Oncol 2016;11:1682-9. [Crossref] [PubMed]
- Chabon JJ, Simmons AD, Lovejoy AF, et al. Circulating tumour DNA profiling reveals heterogeneity of EGFR inhibitor resistance mechanisms in lung cancer patients. Nat Commun 2016;7:11815. [Crossref] [PubMed]
- Ko B, Paucar D, Halmos B. EGFR T790M: revealing the secrets of a gatekeeper. Lung Cancer (Auckl) 2017;8:147-59. [Crossref] [PubMed]
- Wang S, Cang S, Liu D. Third-generation inhibitors targeting EGFR T790M mutation in advanced non-small cell lung cancer. J Hematol Oncol 2016;9:34. [Crossref] [PubMed]
- Diehl F, Schmidt K, Choti MA, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med 2008;14:985-90. [Crossref] [PubMed]
- Reinert T, Schøler LV, Thomsen R, et al. Analysis of circulating tumour DNA to monitor disease burden following colorectal cancer surgery. Gut 2016;65:625-34. [Crossref] [PubMed]
- Jahr S, Hentze H, Englisch S, et al. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res 2001;61:1659-65. [PubMed]
- Perakis S, Auer M, Belic J, et al. Advances in Circulating Tumor DNA Analysis. Adv Clin Chem 2017;80:73-153. [Crossref] [PubMed]
- Garcia-Murillas I, Schiavon G, Weigelt B, et al. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci Transl Med 2015;7:302ra133. [Crossref] [PubMed]
- De Mattos-Arruda L, Weigelt B, Cortes J, et al. Capturing intra-tumor genetic heterogeneity by de novo mutation profiling of circulating cell-free tumor DNA: a proof-of-principle. Ann Oncol 2014;25:1729-35. [Crossref] [PubMed]
- Fenizia F, Pasquale R, Roma C, et al. Measuring tumor mutation burden in non-small cell lung cancer: tissue versus liquid biopsy. Transl Lung Cancer Res 2018;7:668-77. [Crossref] [PubMed]
- Friedlaender A, Nouspikel T, Christinat Y, et al. Tissue-Plasma TMB Comparison and Plasma TMB Monitoring in Patients With Metastatic Non-small Cell Lung Cancer Receiving Immune Checkpoint Inhibitors. Front Oncol 2020;10:142. [Crossref] [PubMed]
- Higuchi R, Fockler C, Dollinger G, Watson R. Kinetic PCR analysis: real-time monitoring of DNA amplification reactions. Biotechnology (N Y) 1993;11:1026-30. [PubMed]
- Newton CR, Graham A, Heptinstall LE, et al. Analysis of any point mutation in DNA. The amplification refractory mutation system (ARMS). Nucleic Acids Res 1989;17:2503-16. [Crossref] [PubMed]
- Arya M, Shergill IS, Williamson M, et al. Basic principles of real-time quantitative PCR. Expert Rev Mol Diagn 2005;5:209-19. [Crossref] [PubMed]
- Olmedillas-López S, García-Arranz M, García-Olmo D. Current and Emerging Applications of Droplet Digital PCR in Oncology. Mol Diagn Ther 2017;21:493-510. [Crossref] [PubMed]
- Zhong Q, Bhattacharya S, Kotsopoulos S, et al. Multiplex digital PCR: breaking the one target per color barrier of quantitative PCR. Lab Chip 2011;11:2167-74. [Crossref] [PubMed]
- Peng Q, Vijaya Satya R, Lewis M, et al. Reducing amplification artifacts in high multiplex amplicon sequencing by using molecular barcodes. BMC Genomics 2015;16:589. [Crossref] [PubMed]
- Nikolaev S, Lemmens L, Koessler T, et al. Circulating tumoral DNA: Preanalytical validation and quality control in a diagnostic laboratory. Anal Biochem 2018;542:34-9. [Crossref] [PubMed]
- Xu C, Gu X, Padmanabhan R, et al. smCounter2: an accurate low-frequency variant caller for targeted sequencing data with unique molecular identifiers. Bioinformatics 2019;35:1299-309. [Crossref] [PubMed]
- Xu Q, Zhu Y, Bai Y, et al. Detection of epidermal growth factor receptor mutation in lung cancer by droplet digital polymerase chain reaction. Onco Targets Ther 2015;8:1533-41. [PubMed]
- Xu C. A review of somatic single nucleotide variant calling algorithms for next-generation sequencing data. Comput Struct Biotechnol J 2018;16:15-24. [Crossref] [PubMed]
- Heeke S, Benzaquen J, Hofman V, et al. Critical Assessment in Routine Clinical Practice of Liquid Biopsy for EGFR Status Testing in Non-Small-Cell Lung Cancer: A Single-Laboratory Experience (LPCE, Nice, France). Clin Lung Cancer 2020;21:56-65.e8. [Crossref] [PubMed]
- Iwama E, Sakai K, Azuma K, et al. Monitoring of somatic mutations in circulating cell-free DNA by digital PCR and next-generation sequencing during afatinib treatment in patients with lung adenocarcinoma positive for EGFR activating mutations. Ann Oncol 2017;28:136-41. [Crossref] [PubMed]
- Li BT, Janku F, Jung B, et al. Ultra-deep next-generation sequencing of plasma cell-free DNA in patients with advanced lung cancers: results from the Actionable Genome Consortium. Ann Oncol 2019;30:597-603. [Crossref] [PubMed]
- Whale AS, Huggett JF, Cowen S, et al. Comparison of microfluidic digital PCR and conventional quantitative PCR for measuring copy number variation. Nucleic Acids Res 2012;40:e82. [Crossref] [PubMed]
- Zhang BO, Xu CW, Shao Y, et al. Comparison of droplet digital PCR and conventional quantitative PCR for measuring EGFR gene mutation. Exp Ther Med 2015;9:1383-8. [Crossref] [PubMed]
- Feng Q, Gai F, Sang Y, et al. A comparison of QuantStudioTM 3D Digital PCR and ARMS-PCR for measuring plasma EGFR T790M mutations of NSCLC patients. Cancer Manag Res 2018;10:115-21. [Crossref] [PubMed]
- Watanabe M, Kawaguchi T, Isa S, et al. Ultra-Sensitive Detection of the Pretreatment EGFR T790M Mutation in Non-Small Cell Lung Cancer Patients with an EGFR-Activating Mutation Using Droplet Digital PCR. Clin Cancer Res 2015;21:3552-60. [Crossref] [PubMed]
- Vannitamby A, Hendry S, Irving L, et al. Novel multiplex droplet digital PCR assay for scoring PD-L1 in non-small cell lung cancer biopsy specimens. Lung Cancer 2019;134:233-7. [Crossref] [PubMed]
- Mencia-Trinchant N, Hu Y, Alas MA, et al. Minimal Residual Disease Monitoring of Acute Myeloid Leukemia by Massively Multiplex Digital PCR in Patients with NPM1 Mutations. J Mol Diagn 2017;19:537-48. [Crossref] [PubMed]
Cite this article as: Nouspikel T. Digging deep or spreading wide?—a comparison of polymerase chain reaction and sequencing approaches in the analysis of circulating tumor DNA. Precis Cancer Med 2020;3:13.

