Targeting EGFR mutations needs to be more precise with co-mutations
Introduction
Treatment of advanced lung cancer with epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (TKI) has evolved over the past 15 years. Conversely to the story of trastuzumab in HER2 positive breast cancer with the target first being identified and then treatment with the drug, the EGFR mutation target was only discovered after an empiric EGFR TKI drug patient response was identified (1). Even subsequent to that, targeting EGFR evolved from general populations to EGFR protein and gene overexpression to now only an EGFR mutation identified from tissue or plasma is an indication for EGFR TKI treatment in lung cancer (2). A next stratification step was the recognition of the clinical difference between EGFR exon 19 deletions and exon 21 L858R mutations. Exon 19 deletions consistently demonstrated a longer progression free (PFS) and overall survival (OS) than exon 21 L858R mutations despite an equivalent response rate (3). A variety of additional EGFR mutations are now identified as sensitive to EGFR TKI targeting most notably exon 20 T790M, a resistant mutation only sensitive to osimertinib (4). Throughout this evolution five different EGFR TKIs now have United States FDA approval and are commercially available. There is some subset suggestion that afatinib may have a survival benefit for exon 19 deletions and dacomitinib may equalize the outcomes for exon 21 L858R mutations (5,6).
Combining anti-VEGFA/VEGFR-2 monoclonal antibodies with an EGFR TKI have shown prolonged PFS compared to the EGFR TKI alone but a lack of proven OS benefit has limited their acceptance into standard clinical use (7-9). Two studies comparing chemotherapy plus gefitinib to gefitinib alone do show an OS benefit. However, in one study the OS of the gefitinib alone arm was a very inferior 17 months and in a Japanese population study OS in both treatment arms, 50.9 months chemotherapy plus gefitinib and 38.8 months gefitinib alone, were superior to other studies (10,11). Most recently the FLAURA trial update favored first line osimertinib alone over a first/second-generation EGFR TKI with a statistically significant OS benefit of 38.6 versus 31.8 months (HR of 0.799 and P=0.0462), yet at 4 years the Kaplan-Meier curves have come together (12).
Case presentation
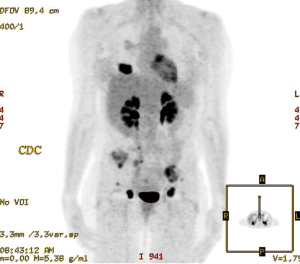
A 57-year-old female never smoker presented with a progressing recalcitrant nonproductive cough and left flank pain. Imaging confirmed a RUL lung mass with mediastinal adenopathy and bone metastases (Figure 1). Transbronchial biopsy of the RUL primary tumor mass with a TTF-1 positive adenocarcinoma.
Tumor management
Plasma next-generation sequencing (NGS) identified an EGFR exon 19 deletion with a mutant allele fraction (MAF) of 10.9% and also a KDR Q472H mutation with a MAF of 11.3% along with two TP53 mutations in exon 5/MAF 4.7% and exon 6/MAF 2.3%, a PTEN mutation MAF 3.8% and a PIK3CA V344A mutation/MAF 3.9%.
Given the KDR Q472H co-mutation, therapy was initiated with combination erlotinib and bevacizumab. A complete response was achieved within 4 months (Figure 2). Treatment with erlotinib and bevacizumab is ongoing.
Case-related literature review
First line treatment decisions in treating advanced EGFR mutated lung cancer remain with group options of an EGFR TKI alone or in combination with chemotherapy or anti-VEGF/VEGFR-2 monoclonal antibodies. However, studies with these options have not been fully compared and there is nothing beyond the EGFR mutation to guide individual treatment decision making. The evolution of treating EGFR mutated lung cancer has been based upon group data. Throughout the evolution of EGFR targeted therapies, what has been lacking in all studies have been the differential impact and potential therapeutic implication of an individual’s co-occurring mutations.
With the evolution of tissue and plasma NGS, a broad array of co-occurring mutations can be identified in the EGFR mutation lung cancer population. Extended molecular testing by either tissue and/or plasma NGS has been well studied to identify and guide treatment of the many secondary EGFR TKI resistance pathways. As treatment advances evolve, so do resistance pathways. With the first/second generation TKIs, exon 20 T790M mutations are most frequently identified. Resistant pathways can be differentially based upon persistence or clearance of the EGFR mutation. With acquired resistance to osimertinib, EGFR C797S mutations are the most frequent resistance escape when the T790M mutation persists, whereas when T790M is lost at resistance, small cell transformation, KRAS mutations and gene fusions occur with a more rapid time to resistance (13).
It is also becoming evident co-occurring mutations in EGFR mutated lung cancer at the time of diagnosis are of prognostic and potentially predictive impact. In addition to their utility in secondary resistance, co-occurring mutations can identify primary EGFR TKI resistance pathways impacting treatment outcomes (14-18). In a large cohort of 200 metastatic EGFR mutated lung cancers, extended pre-treatment molecular testing with versions of the MSK-IMPACT NGS panel identified TP53 mutations in 60% of patients, PIK3CA in 12%, and CTNNB1 in 9% with a median of five co-occurring mutations. ERBB2 amplification was seen in 4% and MET amplification in 2% of patients. Shorter time to progression was seen in the TP53 mutation and ERBB2 and MET amplification patients (19). With broad NGS testing on patients with confirmed primary first/second generation EGFR TKI resistance, the non-driver mutations TP53 P72R, KDR Q472H, and KIT M541L, beyond T790M, were most frequently associated with primary resistance (20). All resistant patients also had other coexisting somatic mutations. Extended genomic profiling of a cohort treated with first/second generation EGFR TKIs identified 50% TP53, 10% PIK3CA and 5% PTEN co-occurring mutations. Median PFS was significantly shorter at 6.5 months compared to TP53 wild type PFS of 19 months and an OS of just 15 months in EGFR exon 19 deletion mutated patients (21).
TP53 is also unfavorably prognostic with poorer survivals in the T790M mutation population. In a larger experience of 131 stage IV EGFR mutated lung cancer patients, 62% had TP53 and 42% other co-occurring mutations. Patients with T790M and TP53 dual mutations had a worse OS than the EGFR T790M and TP53 wild type patients (22). TP53 mutation status can also impact the central nervous system benefit of EGFR TKI treatment. In 100 EGFR mutated patients with brain metastasis treated with osimertinib, a disease control rate of 29% in TP53 mutated patients was far inferior to the 94% achieved in TP53 wild type patients. Additionally, all of the exon 8 TP53 mutations were osimertinib resistant (23).
EGFR treated small cell transformation risk can also be identified with upfront extended molecular testing. Concurrent TP53 with RB1 mutations at diagnosis are prognostically unfavorable and can portend transformation to small cell histology and biology recurrences. In 863 EGFR mutated lung cancers, tissue NGS identified coexisting TP53 and RB1 mutations in 5%. There was a de novo or subsequent small cell transformation recurrence in 25% of the patients with TP53 and RB1 mutations at diagnosis, yet none of the patients without these baseline mutations developed a small cell transformation recurrence (24).
This identification and understanding of the clinical utility of co-occurring mutations in guiding individual treatment approaches in the clinic beyond group single agent EGFR TKI use is evolving. Non-driver co-occurring mutations are clearly unfavorably prognostic. What predictive benefit is still unknown and unstudied.
Uniquely in addition to an association with resistance, a kinase insert domain receptor (KDR) co-mutation may also guide a differing treatment approach than just an EGFR TKI alone. KDR mutations are frequently present in lung adenocarcinomas (25,26). The KDR Q472H mutation has been associated with primary EGFR TKI resistance (20). The KDR gene encodes VEGFR-2 function. KDR Q472H mutations are associated with elevated VEGFA levels and increased tumor microvessel density (27). Preclinical studies confirm VEGF upregulation with a rise in tumor and stromal VEGF resulting in EGFR TKI resistance (28). Dual combined EGFR/VEGF pathway blockade can reverse secondary and overcome primary EGFR TKI resistance (29-31). There is a notable differential response in secondary and primary resistance. The combination of bevacizumab and erlotinib showed stronger inhibition of tumor microvessel density and tumor cell proliferation in the erlotinib sensitive phase whereas after establishment of resistance, there was limited antitumor efficacy (32). Effectiveness of this dual therapy approach extends to T790m mutations (33). This provides strong preclinical support for combining bevacizumab or ramucirumab with an EGFR TKI in the presence of a KDR Q472H co-occurring mutation with a sensitive EGFR mutation.
In this light, three upfront bevacizumab plus erlotinib and now ramucirumab plus erlotinib studies are particularly intriguing. Studies comparing combination EGFR TKI therapy with either the anti-VEGFA monoclonal antibody bevacizumab and most recently the anti-VEGFR-2 monoclonal antibody ramucirumab, have demonstrated significantly improved PFS compared to the TKI alone. The combination bevacizumab trials showed a PFS of 16 months in JO25567 and 18.9 months in NEJ026 significantly better than the EGFR TKI alone (7,8). Any OS benefit remains unclear as follow up is ongoing. Both trials are in Asian populations with a meta-analysis finding an ethnicity difference in populations treated with erlotinib and bevacizumab. The Asian/Pacific Islander subset of patients treated with erlotinib and bevacizumab had the most favorable PFS benefit with a HR of 0.23 (P=0.001) (34). An American trial identified a numerically longer albeit statistically insignificant PFS difference of 17.9 months with the bevacizumab/erlotinib combination versus 13.5 months with erlotinib alone (35).
The combination ramucirumab RELAY trial showed a PFS of 19.4 versus 12.4 months for erlotinib alone (9). OS is premature. Cross trial comparison limitations aside, the RELAY trial comparing ramucirumab and erlotinib versus erlotinib alone and the FLAURA study of first line osimertinib versus either erlotinib or gefitinib, show very similar group results in very similar patient populations. The RELAY study population included 76% Asians with 62% in FLAURA. Response rates were similar with 76% overall and 1% complete responses in RELAY with 80% overall and 3% complete responses in FLAURA. PFS was also strikingly similar at 19.4 months in RELAY and 18.9 months in FLAURA (9,12). Whatever cross trial similarities, what is lacking (or known) in both and all EGFR TKI studies are the co-occurring mutation findings and specific treatment arm benefit or detrimental impact.
The KDR Q472H mutation and VEGFR-2/VEGFA pathway EGFR TKI resistance interconnection with a potential anti-VEGFA/VEGFR-2 benefit when used in the first-line setting, guided the rationale for our treatment decision of utilizing upfront combined bevacizumab with erlotinib in our patient. In our opinion, there is a very compelling preclinical foundation and clinical rationale supporting treatment with an EGFR TKI and anti-VEGFA/VEGFR-2 monoclonal combination in the setting of a KDR Q472H co-occurring mutation. Unfortunately, there is no clinical data to support or refute this treatment approach. At best, data will not always be there, but individual treatment decisions will still need to be made. Group data does not always apply to the individuals within the group. We learn from groups, yet we take care of individuals. Tumor biology is individual and when known, needs to be individually treated. Non-driver mutations are a distinct part of an individual’s tumor biology. Co-occurring mutations are clearly prognostic indicating EGFR TKI resistance. This immediately emphasizes the next question in the treatment of EGFR mutated lung cancer; can certain co-occurring mutations also be predictive of a better treatment approach for each individual than an EGFR TKI alone?
Even a retrospective assessment of stored tissue or plasma NGS co-occurring mutations in completed EGFR treatment studies could well better identify enriched individual subsets best benefiting from what treatment arm, whether TKI alone, or in combination with chemotherapy or anti-VEGFA/VEGFR-2 therapy. Clarifying the need for and timing of radiation therapy in addition to a central nervous system penetrating EGFR TKI in treating brain metastases could be guided by identified co-occurring mutation group subsets. This would not be a post hoc subset manipulation as it would only reflect the tumor biology of each treated individual and their outcome. That is exactly how the EGFR mutation TKI treatment advance came forth and is precisely precision oncology. Certainly, all EGFR mutated lung cancer treatment studies going forward should include upfront broad molecular NGS testing to identify the prognostic and predictive impact of co-occurring mutations.
Conclusions
Certain coexisting non-driver mutations in advanced EGFR mutated lung cancers are adversely prognostic. KDR Q472H mutations are associated with VEGF upregulation and EGFR TKI resistance, including primary resistance. As supported by preclinical data and prolonged PFS in clinical studies, KDR Q472H mutations could well also be predictive of a reversible resistance pathway with the combination of anti-VEGFA/VEGFR-2 monoclonal antibodies and EGFR TKIs. It is imperative to prospectively include co-occurring mutation findings and outcomes in all EGFR mutated lung cancer studies. These non-driver co-occurring mutations do not have to always be ‘targetable’ to be ‘actionable’ guiding different therapeutic approaches for each individual. Trastuzumab would have failed if the amplified HER2 population was blended into the larger non-HER2 selected breast cancer population. The precision oncology treatment of advanced EGFR mutated lung cancers needs to become more precise with the prognostic and predictive inclusion of co-occurring mutations.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the editorial office, Precision Cancer Medicine for the series “Precision Oncology Tumor Board”. The article has undergone external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/pcm.2019.11.06). The Series “Precision Oncology Tumor Board” was commissioned by the editorial office without any funding or sponsorship. PRW reports personal fees from Genentech, Biodesix and Circulogene, outside the submitted work. PRW is now Chief Medical Officer at Circulogene. At the time of this patient care, PRW was not associated with Circulogene, and has no conflicts of interest to declare. PRW serves as an unpaid editorial board member of Precision Cancer Medicine from Feb 2019 to Jan 2021. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee(s) and with the Helsinki Declaration (as revised in 2013). The patient gives her oncologists informed consent for public discussion and de-identified use in medical journals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 2004;350:2129-39. [Crossref] [PubMed]
- Cappuzzo F, Hirsch FR, Rossi E, et al. Epidermal Growth Factor Receptor Gene and Protein and Gefitinib Sensitivity in Non-Small-Cell Lung Cancer. J Natl Cancer Inst 2005;97:643-55. [Crossref] [PubMed]
- Jackman DM, Yeap BY, Sequist LV, et al. Exon 19 Deletion Mutations of Epidermal Growth Factor Receptor Are Associated with Prolonged Survival in Non-Small Cell Lung Cancer Patients Treated with Gefitinib or Erlotinib. Clin Cancer Res 2006;12:3908-14. [Crossref] [PubMed]
- Mok TS, Wu YL, Ahn MJ, et al. Osimertinib or platinum-pemetrexed in EGFR T790M-positive lung cancer. N Engl J Med 2017;376:629-40. [Crossref] [PubMed]
- Yang JC, Wu Y, Schuler M, et al. Afatinib versus cisplatin-based chemotherapy for EGFR mutation-positive lung adenocarcinoma (LUX-Lung 3 and LUX-Lung 6): analysis of overall survival data from two randomized phase 3 trials. Lancet Oncol 2015;16:141-51. [Crossref] [PubMed]
- Mok TS, Cheng Y, Zhou X, et al. Improvement in Overall Survival in a Randomized Study That Compared Dacomitinib With Gefitinib in Patients With Advanced Non-Small-Cell Lung Cancer and EGFR-Activating Mutations. J Clin Oncol 2018;36:2244-50. [Crossref] [PubMed]
- Saito H, Fukuhara T, Furuya N, et al. Erlotinib plus bevacizumab versus erlotinib alone in patients with EGFR-positive advanced non-squamous non-small-cell lung cancer (NEJ026): interim analysis of an open-label, randomized, multicenter, phase 3 trial. Lancet Oncol 2019;20:625-35. [Crossref] [PubMed]
- Yamamoto N, Seto T, Nishio M, et al. Erlotinib plus bevacizumab (EB) versus erlotinib € alone as first-line treatment for advanced EGFR-mutation-positive non-squamous non-small-cell lung cancer (NSCLC): Survival follow-up results of JO25567. J Clin Oncol 2018;36:
- Nakagawa K, Garon EB, Seto T, et al. Ramucirumab plus erlotinib in patients with untreated, EGFR-mutated, advanced non-small-cell lung cancer (RELAY): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol 2019; [Epub ahead of print]. [Crossref] [PubMed]
- Noronha V, Patil VM, Joshi A, et al. Gefitinib Versus Gefitinib Plus Pemetrexed and Carboplatin Chemotherapy in EGFR-Mutated Lung Cancer. J Clin Oncol 2019; [Epub ahead of print]. [PubMed]
- Hosomi Y, Morita S, Sugawara S, et al. Gefitinib Alone Versus Gefitinib Plus Chemotherapy for Non-Small-Cell Lung Cancer With Mutated Epidermal Growth Factor Receptors: NEJ009 Study. J Clin Oncol 2019; [Epub ahead of print]. [PubMed]
- Ramalingam SS, Gray JE, Ohe Y, et al. Osimertinib vs comparator EGFR-TKI as first-line treatment for EGFRm advanced NSCLC (FLAURA): Final overall survival analysis. Ann Oncol 2019;30:v851-934. [Crossref]
- Oxnard GR, Hu Y, Mileham KF, et al. Assessment of Resistance Mechanisms and Clinical Implications in Patients With EGFR T790M-Positive Lung Cancer and Acquired Resistance to Osimertinib. JAMA Oncol 2018;4:1527-34. [Crossref] [PubMed]
- Skoulidis F, Heymach JV. Co-occurring genomic alterations in non-small-cell lung cancer biology and therapy. Nat Rev Cancer 2019;19:495-509. [Crossref] [PubMed]
- Blakely CM, Watkins TBK, Wu W, et al. Evolution and clinical impact of co-occurring genetic alterations in advanced-stage EGFR-mutant lung cancers. Nat Genet 2017;49:1693-704. [Crossref] [PubMed]
- Rachiglio AM, Fenizia F, Piccirillo MC, et al. The Presence of Concomitant Mutations Affects the Activity of EGFR Tyrosine Kinase Inhibitors in EGFR-Mutant Non-Small Cell Lung Cancer (NSCLC) Patients. Cancers 2019; [Crossref] [PubMed]
- Chen M, Xu Y, Zhao J, et al. Concurrent Gene Mutations as Negative Predictive Factors in Epidermal Growth Factor Receptor-Positive non-Small Cell Lung Cancer. EBioMedicine 2019;42:304-10. [Crossref] [PubMed]
- Wang Z, Cheng Y, An T, et al. Detection of EGFR mutations in plasma circulating tumour DNA as a selection criterion for first-line gefitinib treatment in patients with advanced lung adenocarcinoma (BENEFIT): a phase 2, single-arm, multicenter clinical trial. Lancet Respir Med 2018;6:681-90. [Crossref] [PubMed]
- Yu HA, Suzawa K, Jordan E, et al. Concurrent alterations in EGFR-mutant lung cancer associated with resistance to EGFR kinase inhibitors and characterization of MTOR as a mediator of resistance. Clin Cancer Res 2018;24:3108-18. [Crossref] [PubMed]
- Masago K, Fujita S, Muraki M, et al. Next-generation sequencing of tyrosine kinase inhibitor-resistant non-small-cell lung cancers in patients harboring epidermal growth-factor-activating mutations. BMC Cancer 2015;15:908-15. [Crossref] [PubMed]
- VanderLaan PA, Rangachari D, Mockus AM, et al. Mutations in TP53, PIK3CA, PTEN and other genes in EGFR mutated lung cancers: Correlation with clinical outcomes. Lung Cancer 2017;106:17-21. [Crossref] [PubMed]
- Aggarwal C, Davis CW, Mick R, et al. Influence of TP53 Mutation on Survival in Patients With Advanced EGFR-Mutant Non-Small-Cell Lung Cancer. JCO Precis Oncol 2018. doi:
10.1200/PO.18.00107 . - Chen L, Mu X, Wu H, et al. Association between TP53 mutations and efficacy of Osimertinib for brain metastasis from EGFR-mutant lung cancer. Ann Oncol 2019;30:v143-8.
- Offin M, Chan JM, Tenet M, et al. Concurrent RB1 and TP53 Alterations Define a Subset of EGFR-Mutant Lung Cancers at risk for Histologic Transformation and Inferior Clinical Outcomes. J Thorac Oncol 2019;14:1784-93. [Crossref] [PubMed]
- Ding L, Getz G, Wheeler DA, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature 2008;455:1069-75. [Crossref] [PubMed]
- Mockus SM, Potter CS, Stafford GA, et al. Targeting KDR mutations in lung adenocarcinoma. Cancer Res 2015;75:73.
- Glubb DM, Cerri E, Giese A, et al. Novel Functional Germline Variants in the VEGF Receptor 2 Gene and Their Effect on Gene Expression and Microvessel Density in Lung Cancer. Clin Cancer Res 2011;17:5257-67. [Crossref] [PubMed]
- Viloria-Petit A, Crombet T, Jothy S, et al. Acquired Resistance to the Antitumor Effect of Epidermal Growth Factor Receptor-blocking Antibodies in Vivo: A Role for Altered Tumor Angiogenesis. Cancer Res 2001;61:5090-101. [PubMed]
- Naumov GN, Nilsson MB, Cascone T, et al. Combined Vascular Endothelial Growth Factor Receptor and Epidermal Growth Factor Receptor (EGFR) Blockade Inhibits Tumor Growth in Xenograft Models of EGFR Inhibitor Resistance. Clin Cancer Res 2009;15:3484-94. [Crossref] [PubMed]
- Watanabe H, Ichihara E, Kayatani H, et al. Significant combination benefit of anti-VEGFR antibody and oncogene-targeted agents in EGFR or ALK mutant NSCLC cells. AACR Annual Meeting 2019;abstract 2131.
- Li H, Takayama K, Wang S, et al. Addition of bevacizumab enhances antitumor activity of erlotinib against non-small cell lung cancer xenografts depending on VEGF expression. Cancer Chemother Pharmacol 2014;74:1297-305. [Crossref] [PubMed]
- Masuda C, Yanagisawa M, Yorozu K, et al. Bevacizumab counteracts VEGF-dependent resistance to erlotinib in an EGFR-mutated NSCLC xenograft model. Int J Oncol 2017;51:425-34. [Crossref] [PubMed]
- Rosell R, Dafni U, Felip E, et al. Erlotinib and bevacizumab in patients with advanced non-small-cell lung cancer and activating EGFR mutations (BELIEF): an international, multicenter, single-arm, phase 2 trial. Lancet Respir Med 2017;5:435-44. [Crossref] [PubMed]
- Zhang S, Mao X, Wang H, et al. Efficacy and safety of bevacizumab plus erlotinib versus bevacizumab or erlotinib alone in the treatment of non-small-cell lung cancer: a systemic review and meta-analysis. BMJ Open 2016;6:e011714. [Crossref] [PubMed]
- Stinchcombe TE, Janne PA, Wang X, et al. Effect of Erlotinib Plus Bevacizumab vs Erlotinib Alone on Progression-Free Survival in Patients With Advanced EGFR-Mutant Non-Small Cell Lung Cancer. JAMA Oncol 2019;5:1448-55. [Crossref] [PubMed]
Cite this article as: Walker PR, Sharma N, Namireddy P. Targeting EGFR mutations needs to be more precise with co-mutations. Precis Cancer Med 2020;3:8.