
Precision Oncology Tumor Board—a new series to facilitate a global dialogue
Over the last two decades since the first breakthrough discoveries, such as the use of imatinib for chronic myelogenous leukemia (CML) and gastrointestinal stromal tumor (GIST) sarcomas and trastuzumab for ErbB2-positive breast cancers there has been an explosion of knowledge as to our understanding of the molecular genetics of malignancies (1-3). The ready actionability of recurrent molecular alterations has led to a series of pivotal clinical studies establishing molecular testing to guide the management of many advanced malignancies yielding hope that ultimately genomic testing could allow accurate matching of targeted therapies driving improved outcomes across the board. Now 20 years past these initial sentinel events, tens of thousands of cancer genomes have been carefully mapped out through vast efforts such as The Cancer Genome Atlas (TCGA)/International Cancer Genome Consortium (ICGC)/pediatric Cancer Genome Project (CGP) and many others (4). An array of molecularly focused basket and umbrella studies demonstrating a spectrum of benefits have been completed and our treatment focus unexpectedly but with renewed excitement has shifted towards tumor immunology—in essence, the epitome of molecular targeting. So today we are facing a scenario that is certainly more complex and layered and maybe less easily fulfilling than originally anticipated (5,6). However, it is a scenario that provides possibly even more hope than ever as to advancing the treatment of patients suffering from cancer through the best use of a dizzying range of biomarkers now available to the practicing clinician. The need indeed has never been more pressing for sounding boards such as Precision Oncology Tumor Boards to provide proper education and a forum for discussion about the optimal use of biomarker testing in a vast number of emerging clinical settings. It is for this very reason- to provide a global forum for dialogue and education—that we decided to establish a recurring Precision Oncology Tumor Board series.
As a brief introduction to this new series, let us review what we mean by precision oncology as a framework of clinical practice and research. The original discoveries of key actionable, “druggable” oncogenes set up an original school of thought of one gene- one drug associations, an idea yielding tremendous benefits in the molecular classification and management of advanced GIST, breast cancer, melanoma and non-small cell lung cancer amongst others (6). Indeed, molecular oncology completely transformed the landscape of advanced non-small cell lung cancer management where upfront testing for EGFR/ALK/ROS aberrations now is a must, closely followed by up and coming validated and emerging markers, such as B-Raf/MET/NTRK/RET and ErbB2 amongst others (7). These molecular subsets have opened the door for highly effective targeting by an emerging tapestry of generations of potent targeted drugs in fact making non-small cell lung cancer the poster child of molecular oncology. The case of colorectal carcinoma provides a very different facet where extended Ras testing can instruct the clinician to avoid ineffective, costly and somewhat toxic EGFR-targeting drugs for patients harboring alterations downstream of the targeted pathway—providing value-based optimal treatment selection, an equally important use of molecular testing (8). Lastly, molecular testing quickly emerged as a key platform to provide insights into frequently molecularly definable acquired resistance alterations when using effective targeted therapies leading to the rapid emergence of powerful circulating tumor DNA (ctDNA)-based dynamic testing (9) and novel therapies addressing common resistance mechanisms, such as for example EGFR T790M (10,11).
This initial flurry of discoveries led to a series of well-designed biomarker-driven studies yielding many great successes and some sobering failures of the one gene-one target era quickly reaching some level of saturation of checking out the low-hanging fruits of molecular oncology. The low frequency and cross-tumor distribution of many of the remaining alterations called for a series of powerful and novel designs such as basket and umbrella studies now starting to yield real dividends with primary endpoints having been reached in several Molecular Analysis for Therapy Choice (MATCH) and Targeted Agent and Profiling Utilization Registry (TAPUR) study arms while outstanding molecular matching rates have been reported from Pediatric MATCH recently (12-14). Tumor-specific basket studies such as Lung Master Protocol (MAP), Beat acute myeloid leukemia (AML) provide ongoing opportunities for expanding molecular and now biomarker-driven immune oncology-based treatment arms (15,16) and adventurous efforts such as SHIVA, MOSCATO, IMPACT, I-PREDICT and WINTHER show the way as to molecular testing-based cohort studies demonstrating feasibility and initial promise of DNA, ctDNA and RNA-based testing and matching approaches (17-21). However, these studies also highlight some shortcomings of such approaches- uncommon alterations and limited number of available of drugs for now have set a cap on the overall success noted. In addition, beyond the first set of highly actionable alterations many more common recurring events have been a challenge to target, e.g., K-Ras/PIK3CA with some promising leads finally emerging even for these difficult to tackle molecular alterations (22,23). Indeed, very quickly the focus is also shifting towards biomarker-selection for neoadjuvant/curative approaches with the tremendous promise of early read-outs as well as possibly more definitive benefits.
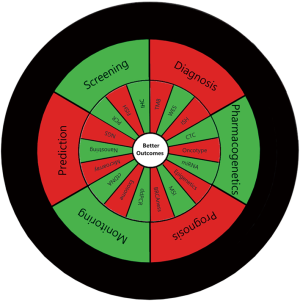
While some saturation as to what can be achieved with molecularly targeted therapy has been noted, we should highlight the rapidly expanding horizon as to what actually should be considered molecular targeting. In addition to addressing drug sensitivities, now molecular testing can many times inform us as to primary and even more effectively acquired resistance mechanisms. Multigene panels and next generation sequencing approaches have opened our eyes onto critically important issues as to clonality and tumor heterogeneity (24,25). We are starting to recognize more complex molecular pathway signatures such as “BRCAness” and treatment associations with synthetic lethality (26,27). Lastly, our initial forays into immunotherapy have led to the rapid recognition of the need for a much more refined understanding of the tumor genome as a potential target for T cell recognition with the emergence of tumor mutation burden, specific mutational subsets, e.g., STK11, DNA repair defects such as MSI/POLE-D/E deficiency and specific neoantigen discovery—ultimately possibly contributing to further enhancement of cellular therapies, such as chimeric antibody receptor engineered T cell (CAR-T) and T cell receptor (TCR) approaches (28-32). Artificial intelligence is anticipated to make a major impact as well in the processing and analyses of the vast amount of complex information from our molecular biomarker assays. Of course, as we consider the great progress we have made in molecular discoveries leading to more effective treatment targeting, we would be amiss not to recognize the similarly powerful achievements noted in other areas—prognostication (e.g., Oncotype), molecular staging (HPV, ErbB2), treatment monitoring [ctDNA, circulating tumor cells (CTC) analyses], early detection (ctDNA) etc. (33-36).
The vast knowledge—accumulated and emerging—calls for a very strong investment in rapid and seamless information exchange and novel educational platforms to ensure that the requisite information as to optimized individual patient-level decisions is in fact readily available for the treating clinician (Figure 1). This particular educational niche is what our introductory series is hoping to help fill. In our view, the role of the Precision Oncology Tumor Board is to educate our global audience not just about particular biomarkers and molecularly targeted treatment approaches but hopes to help demonstrate effective ways to gather the necessary knowledge to enable the clinician to arrive at optimal treatment decisions when facing unique molecular findings. Therefore, our goal is to solicit submissions for this series consisting of actual cases that illustrate a particular scenario where molecular biomarkers can be utilized to optimize patient care. While the case or case series should be the foundation of the manuscripts, we anticipate authors will showcase then the best use of the highlighted biomarker with a concise but relevant review of the literature with appropriate highlighting of the level of evidence supporting key decisions and the informed selection from available treatment or decision options.
We launch this case series with the tremendous hope that it will be instructive and at the same time enjoyable to our readers—and even more importantly hoping that the communication exchange will ultimately fulfil the real promise also leading to optimized treatment to patients. Well, isn’t that what any good Tumor Board should be about? Precisely!
Acknowledgments
The author wishes to express his gratitude for Daniel Halmos’ assistance with figure graphics.
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the editorial office, Precision Cancer Medicine for the series “Precision Oncology Tumor Board”. The article did not undergo external peer review.
Conflicts of Interest: The author has completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/pcm.2019.07.01). The Series “Precision Oncology Tumor Board” was commissioned by the editorial office without any funding or sponsorship. BH serves as the unpaid Guest Editor of the series and an unpaid editorial board member of Precision Cancer Medicine from Apr 2019 to Mar 2021. The author has no other conflicts of interest to declare.
Ethical Statement: The author is accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Druker BJ, Tamura S, Buchdunger E, et al. Effects of a selective inhibitor of the Abl tyrosine kinase on the growth of Bcr-Abl positive cells. Nat Med 1996;2:561-6. [Crossref] [PubMed]
- Joensuu H, Roberts PJ, Sarlomo-Rikala M, et al. Effect of the tyrosine kinase inhibitor STI571 in a patient with a metastatic gastrointestinal stromal tumor. N Engl J Med 2001;344:1052-6. [Crossref] [PubMed]
- Slamon DJ, Leyland-Jones B, Shak S, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med 2001;344:783-92. [Crossref] [PubMed]
- Zhang J, Baran J, Cros A, et al. International Cancer Genome Consortium Data Portal--a one-stop shop for cancer genomics data. Database (Oxford) 2011;2011:bar026. [Crossref] [PubMed]
- Duffy MJ. The war on cancer: are we winning? Tumour Biol 2013;34:1275-84. [Crossref] [PubMed]
- Postel-Vinay S, Boursin Y, Massard C, et al. Seeking the driver in tumours with apparent normal molecular profile on comparative genomic hybridization and targeted gene panel sequencing: what is the added value of whole exome sequencing? Ann Oncol 2016;27:344-52. [Crossref] [PubMed]
- Sandler JE, Kaumaya M, Halmos B. Biomarker use in lung cancer management: expanding horizons. Biomark Med 2018;12:315-20. [Crossref] [PubMed]
- Loree JM, Kopetz S. Recent developments in the treatment of metastatic colorectal cancer. Ther Adv Med Oncol 2017;9:551-64. [Crossref] [PubMed]
- Merker JD, Oxnard GR, Compton C, et al. Circulating Tumor DNA Analysis in Patients With Cancer: American Society of Clinical Oncology and College of American Pathologists Joint Review. J Clin Oncol 2018;36:1631-41. [Crossref] [PubMed]
- Kobayashi S, Boggon TJ, Dayaram T, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med 2005;352:786-92. [Crossref] [PubMed]
- Attarian S, Rahman N, Halmos B. Emerging uses of biomarkers in lung cancer management: molecular mechanisms of resistance. Ann Transl Med 2017;5:377. [Crossref] [PubMed]
- Mullard A. NCI-MATCH trial pushes cancer umbrella trial paradigm. Nat Rev Drug Discov 2015;14:513-5. [Crossref] [PubMed]
- Mangat PK, Halabi S, Bruinooge SS, et al. Rationale and Design of the Targeted Agent and Profiling Utilization Registry (TAPUR) Study. JCO Precis Oncol 2018. doi:
10.1200/PO.18.00122 . - Allen CE, Laetsch TW, Mody R, et al. Target and Agent Prioritization for the Children's Oncology Group-National Cancer Institute Pediatric MATCH Trial. J Natl Cancer Inst 2017; [Crossref] [PubMed]
- Herbst RS, Gandara DR, Hirsch FR, et al. Lung Master Protocol (Lung-MAP)-A Biomarker-Driven Protocol for Accelerating Development of Therapies for Squamous Cell Lung Cancer: SWOG S1400. Clin Cancer Res 2015;21:1514-24. [Crossref] [PubMed]
- Personalized Medicine Applied to AML. Cancer Discov 2019;9:156-7.
- Le Tourneau C, Delord JP, Goncalves A, et al. Molecularly targeted therapy based on tumour molecular profiling versus conventional therapy for advanced cancer (SHIVA): a multicentre, open-label, proof-of-concept, randomised, controlled phase 2 trial. Lancet Oncol 2015;16:1324-34. [Crossref] [PubMed]
- Massard C, Michiels S, Ferte C, et al. High-Throughput Genomics and Clinical Outcome in Hard-to-Treat Advanced Cancers: Results of the MOSCATO 01 Trial. Cancer Discov 2017;7:586-95. [Crossref] [PubMed]
- Tsimberidou AM, Hong DS, Ye Y, et al. Initiative for Molecular Profiling and Advanced Cancer Therapy (IMPACT): An MD Anderson Precision Medicine Study. JCO Precis Oncol 2017. doi:
10.1200/PO.17.00002 . - Sicklick JK, Kato S, Okamura R, et al. Molecular profiling of cancer patients enables personalized combination therapy: the I-PREDICT study. Nat Med 2019;25:744-50. [Crossref] [PubMed]
- Rodon J, Soria JC, Berger R, et al. Genomic and transcriptomic profiling expands precision cancer medicine: the WINTHER trial. Nat Med 2019;25:751-8. [Crossref] [PubMed]
- Ni D, Li X, He X, et al. Drugging K-Ras(G12C) through covalent inhibitors: Mission possible? Pharmacol Ther 2019; [Epub ahead of print]. [Crossref] [PubMed]
- Andre F, Ciruelos E, Rubovszky G, et al. Alpelisib for PIK3CA-Mutated, Hormone Receptor-Positive Advanced Breast Cancer. N Engl J Med 2019;380:1929-40. [Crossref] [PubMed]
- Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 2012;366:883-92. [Crossref] [PubMed]
- Jamal-Hanjani M, Wilson GA, McGranahan N, et al. Tracking the Evolution of Non-Small-Cell Lung Cancer. N Engl J Med 2017;376:2109-21. [Crossref] [PubMed]
- Lord CJ, Ashworth A. BRCAness revisited. Nat Rev Cancer 2016;16:110-20. [Crossref] [PubMed]
- Severson TM, Wolf DM, Yau C, et al. The BRCA1ness signature is associated significantly with response to PARP inhibitor treatment versus control in the I-SPY 2 randomized neoadjuvant setting. Breast Cancer Res 2017;19:99. [Crossref] [PubMed]
- Kitajima S, Ivanova E, Guo S, et al. Suppression of STING Associated with LKB1 Loss in KRAS-Driven Lung Cancer. Cancer Discov 2019;9:34-45. [Crossref] [PubMed]
- Conway JR, Kofman E, Mo SS, et al. Genomics of response to immune checkpoint therapies for cancer: implications for precision medicine. Genome Med 2018;10:93. [Crossref] [PubMed]
- Campbell BB, Light N, Fabrizio D, et al. Comprehensive Analysis of Hypermutation in Human Cancer. Cell 2017;171:1042-1056.e10. [Crossref] [PubMed]
- Lee CH, Yelensky R, Jooss K, et al. Update on Tumor Neoantigens and Their Utility: Why It Is Good to Be Different. Trends Immunol 2018;39:536-48. [Crossref] [PubMed]
- Le DT, Uram JN, Wang H, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med 2015;372:2509-20. [Crossref] [PubMed]
- Sparano JA, Gray RJ, Makower DF, et al. Adjuvant Chemotherapy Guided by a 21-Gene Expression Assay in Breast Cancer. N Engl J Med 2018;379:111-21. [Crossref] [PubMed]
- Leemans CR, Snijders PJF, Brakenhoff RH. The molecular landscape of head and neck cancer. Nat Rev Cancer 2018;18:269-82. Erratum in: Publisher Correction: The molecular landscape of head and neck cancer [Nat Rev Cancer 2018]. [Crossref] [PubMed]
- Chaudhuri AA, Chabon JJ, Lovejoy AF, et al. Early Detection of Molecular Residual Disease in Localized Lung Cancer by Circulating Tumor DNA Profiling. Cancer Discov 2017;7:1394-403. [Crossref] [PubMed]
- Moding EJ, Diehn M, Wakelee HA. Circulating tumor DNA testing in advanced non-small cell lung cancer. Lung Cancer 2018;119:42-7. [Crossref] [PubMed]
Cite this article as: Halmos B. Precision Oncology Tumor Board—a new series to facilitate a global dialogue. Precis Cancer Med 2019;2:23.

