
Alternative non-homologous end joining repair: a master regulator of genomic instability in cancer
Introduction
Genomic instability—a cancer hallmark
Genomic instability refers to biological events that confer a tendency to accumulate genetic defects within the genome. By promoting deleterious mutations, genomic instability progressively enables the acquisition of hallmarks that drive tumor onset and progression (1,2). Several types of genomic instability are described: nucleotide instability (NIN), characterized by base deletions, insertions or substitutions (3); microsatellite instability (MSI) characterized by the expansion or contraction of oligonucleotide repeats present in microsatellite regions (4,5); chromosomal instability, which is the most represented type of genomic instability, characterized by numeric or structural chromosomal variations (6,7).
Evaluation of genomic instability, such as the presence of certain types of translocations or deletions, has acquired a consolidated prognostic value in cancer patients (8,9). More recently, it has been demonstrated that it represents also an important predictive factor, given the opportunity to selectively target a cancer cell with a synthetic lethality approach (10) in the case of homologous recombination (HR) deficiency or with PD1 blockade in the case of MSI (11). So, efforts to unravel the underlying mechanisms which lead to genomic instability is a challenge to overcome for the full exploitation of precision oncology.
DNA repair pathways: an overview
Although molecular basis of genomic instability is largely unknown, growing evidence suggests that perturbation of DNA repair machinery could play a critical role, since it permits to tolerate DNA damage overload, thus selecting cells with advantage in proliferation and survival genes (12).
Human cells are continuously injured by a large amount (approximately 70,000/day) of DNA lesions (13). DNA damage can derive from endogenous sources as oxidative damage, replication fork collapse and telomere erosion, as well from exogenous agents like ionizing radiation (IR) or UV light or chemical exposure. The majority of lesions are single-strand DNA (ssDNA) breaks, which can be randomly converted to more dangerous DNA double-strand breaks (DSBs) (14). However, DSBs are generated also as programmed process in physiological events such V(D)J recombination and class switch recombination (CSR), which occur during immunoglobulin diversity and function generation.
To preserve genomic stability, several DNA repair pathways work together to avoid dangerous effects of unrepaired DSBs. Indeed, if DSBs are too extensive, normal cells activate apoptosis program to prevent propagation of damaged progeny. Otherwise, in cancer cells DSB are often repaired erroneously, leading to genomic instability which could drive oncogenic transformation and progression.
The two major pathways involved in DSB repair are classical non-homologous end joining (C-NHEJ) and HR.
C-NHEJ can occur in all phases of the cell cycle and therefore represents the major pathway for the repair of DSBs (15). C-NHEJ repair is end resection-independent and is a very fast process. DSB recognition is operated by the Ku70-Ku80 heterodimer which binds to DNA
ends and then recruits the DNA-dependent protein kinase catalytic subunit (DNA-PKcs) to phosphorylate Artemis that in turn processes single-stranded overhangs. Finally DNA ligase 4 (LIG4) and the scaffold protein XRCC4 catalyze the ligation of DNA ends. Although C-NHEJ repairs DSBs without a homologous DNA template, its high efficiency associated with only limited sequence alterations at the junction render this pathway a guardian of genomic stability (16).
In contrast to C-NHEJ, HR operates during S and G2 phases of the cell cycle since it requires a homologous template to faithfully restore the sequence around the DSB (17). The first step is represented by DNA end resection in which terminal nucleotides in the 5’ ends are digested generating long 3’ single-stranded DNA (ssDNA) overhangs. This process is operated by Mre11, Rad50, Nbs1 (MRN complex) and accessory proteins like CtIP and BRCA1, which sense the DSBs and initiate recruitment of HR machinery to DSB foci. The 3’-ssDNA tails are then coated and stabilized by the Replication protein A (RPA) complex and Rad51 nucleoprotein filament, followed by strand invasion into the adjacent intact sister chromatid and D-loop formation. Finally, a polymerase catalyzes DNA synthesis until the Holliday junctions become resolved and DSB repaired (18).
In addition to C-NHEJ and HR, recent experimental evidence demonstrates the existence of a third DSB repair pathway named alternative non-homologous end joining (Alt-NHEJ) (19-21).
In this review we focus on the role of Alt-NHEJ repair in the promotion of genomic instability, highlighting its potential value as innovative therapeutic target for personalized treatments.
Alt-NHEJ de-regulation in cancer
Alt-NHEJ machinery
Alt-NHEJ repair comprises three sub-pathways that differentially operate on the basis of DNA sequence complementarity degree at DNA ends of DSBs. Microhomology-mediated end joining (MMEJ) requires from 2 to 20 nucleotides of sequence homology (21); single strand annealing (SSA) operates with homology sequences of more than 25 nucleotides; finally, a third poorly characterized end joining (EJ) pathway which involves very little sequence homology at the repair site.
We will refer to MMEJ to describe Alt-NHEJ, since it is the better characterized subpathway in the aim of our review (Figure 1).
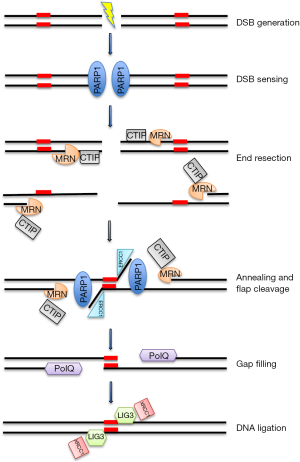
Although is most active in the S and G2 phases of the cell cycle, Alt-NHEJ repair of DSBs also occurs in G1 cells (22). The first step requires DNA End Resection by MRN/CtlP complex (23), specifically recruited to DSB by poly (ADP ribose) polymerase-1 (PARP-1), which acts as sensor for DNA discontinuities (24). In particular, CtIP activation enhances the MRN endonuclease/exonuclease activity (25) resulting in exposition of microhomology sequence within single strand regions. The next step is the bridging and alignment of the DNA ends via the short microhomologies, operated in conjunction by PARP-1, MRN and PolQ activity (26,27). Then, Non-homologous 3’ tails are digested by ERCC1\XPF nucleases generating gaps within DNA strands, which are filled by PolQ-mediated DNA synthesis (27). In the last step, DNA DSBs are finally repaired by DNA ligase 3 (LIG3)/XRCC1 complex. In particular, the scaffold protein XRCC1 drives LIG3 in proximity of DNA breaks by physical interaction with PARP-1 and MRN complex, to finally join DSB (28,29). Although, LIG3 is considered more effective for the final step of DNA ligation, DNA ligase 1 (LIG1) could also play this function in the absence of LIG3 (30).
Relationship with other DSB repair pathways
Several studies propose the role of Alt-NHEJ as back-up pathway when C-NHEJ or HR are defective (31) (Figure 2). Although different mechanisms have been suggested, it is possible that Alt-NHEJ takes over in DSB repair when C-NHEJ or HR machinery have attempted DNA ends processing but somehow failed during the course of repair:
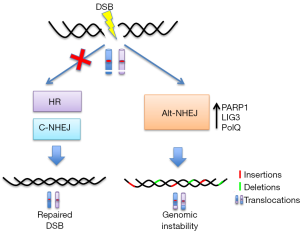
- DSB sensing: DNA breaks sensors PARP-1 and Ku could compete for repair of DSB (32) and the higher affinity of Ku for ends could explain the predominance of C-NHEJ. Indeed down-regulation of Ku leds to increase of Alt-NHEJ activity;
- DNA end processing: CtIP phosphorylation by CDKs (33) or de-acetylation by SIRT6 enhances Alt-NHEJ (34), while 53BP1 blocks CtIP from accessing DNA ends directing repair through C-NHEJ (22);
- gap filling: PolQ has a RAD51-binding domain that inhibits HR thereby promoting ALT-NHEJ (35); moreover Polθ-helicase activity facilitates the removal of RPA from resected DSBs to allow their annealing and subsequent joining by Alt-NHEJ (36);
- DNA ligation: DNA end joining repair in LIG4-null cells is compensated by LIG3-driven Alt-NHEJ (30).
Beyond the role of backup pathway other physiological roles for Alt-NHEJ have been also hypothesized. Indeed, a recent study suggests that Alt-NHEJ is activated upon certain forms of DSBs with damaged termini, like those induced by IR (37). Furthermore, Alt-NHEJ repair play a pivotal role on DSB repair of mitochondrial DNA (mtDNA), since LIG3 is the principal DNA ligase of mitochondria (38).
Alt-NHEJ and genomic instability
Alt-NHEJ is considered an error prone DNA repair pathway for several reasons: (I) Impossibility to correctly restore original DNA sequence in the proximity of DSB due to absence of template strand (31); (II) non-accurate end-joining mechanisms characterized by promiscuity in utilized factors and in delayed repair kinetics (19); (III) low fidelity gap filling by PolQ, which generates significant sequence aberration (insertions) at repair foci, due to its terminal transferase activity and iterative synthesis of ssDNA (39,40); (IV) large deletions produced by endonuclease/exonuclease machinery to unmask microhomologies regions (41); (V) joining of unrelated DNA molecules, due to the N-terminal zinc finger domain (ZnF) of LIG3 (42), which is responsible for generating translocations (43). These genomic aberrations are often caused by errors during V(D)J and CSR recombination, in which programmed DSB are predominantly rejoined by the C-NHEJ pathway, a suppressor of genomic instability (44-46). Consistently, C-NHEJ-deficient mice (due to the lack of Ku80, XRCC4, ligase IV, DNA-PKcs, or even Artemis) that are also defective for p53, often develop pro-B cell lymphomas harboring oncogenic chromosomal translocations involving the IgH and c-Myc loci (47), all of which are catalyzed by Alt-NHEJ. Moreover, SCID mice, which are defective in any of C-NHEJ proteins (48,49), frequently develop tumors with translocations involving the Ig locus generated by Alt-NHEJ. In contrast, mouse cells deficient for Alt-NHEJ core components PARP-1 and LIG3 or PARP inhibitor treated cells exhibit a reduced overall frequency of chromosomal translocations and so, a lower level of genomic instability (43,50).
Alt-NHEJ inhibition strategies
Alt-NHEJ represents a promising therapeutic target in HR or NHEJ defective cancer, since it mainly cooperates as back-up pathway for this two major DSB repair pathways. Indeed, inhibition of Alt-NHEJ should kill only cancer cells that are dependent from Alt-NHEJ pathway to repair their DNA damage sparing normal cells.
Alt-NHEJ inhibition can be currently achieved by blocking: (I) early process of DNA sensing by PARP-1/2 inhibitor; (II) final step of DNA end-joining by DNA ligases LIG1/LIG3 inhibitors:
- PARP1 is the best-characterized member of the PARP family. It senses DNA damage via its DNA binding domain, subsequently synthesizes poly (ADP-ribose) (PAR) which is added to itself and other acceptor proteins. Resulting PARylation recruits other DNA repair proteins including LIG3 and XRCC1 (51,52).
- PARP inhibitors exerts their activity by competing with NAD+ for the catalytic subunit of PARP (53). The evidence that PARP inhibitors induce synthetic lethality cell death in HR deficient cancer cells (54,55) is supported by different mechanism of action (56). Since PARP1 is involved in BER-mediated repair, the prevalent model suggests that DNA SSBs arising during normal cellular activity, persist upon PARP inhibitor treatment and are converted to DSBs which induce cell death in HR defective cells. However, the absence of SSB accumulation during PARP inhibitor treatment leads to hypothesize an alternative explanation for synthetic lethality observed in HR deficient cancer cells treated with PARP-inhibitors (57). In this regard, an intriguing model suggest that HR-defective cells are highly dependent from Alt-NHEJ backup pathway, which is blocked by PARP-inhibitors leading to DNA damage overload and apoptotic cell death (35).
- Consistently with this preclinical evidence, Food and Drug Administration (FDA) has approved three PARPi (olaparib, rucaparib, and niraparib) for the treatment of ovarian cancer (especially relapsed platinum-sensitive high-grade serous disease including those with gBRCAm), and olaparib for treatment of gBRCAm breast cancer.
- However, several challenges will need to be overcome including the identification of predictive biomarkers, such as HR repair deficiency signatures, to precisely identify patient subsets who may have benefit from PARPi alone or combination with chemotherapy, targeted agents, radiotherapy, or immunotherapy;
- DNA ligases are ATP-dependent enzymes which catalyze end-joining of DNA strands. The catalytic core adopts an extended conformation in the absence of DNA, while it forms ring-shaped structures around nicked DNA in presence of DNA damage. LIG3 differs from LIG1 and LIG4 in different ways. First, alternative translation initiation of LIG3 gene generates two isoforms (58) with different cellular functions and localization: (a) nuclear LIG3 which operates in excision repair process and Alt-NHEJ repair (59); (b) mitochondrial LIG3 which is involved in mitochondrial DNA metabolism (38,60). Second, LIG3 have a specific N-terminal ZnF which plays a critical role both in the sensing of DNA breaks and intermolecular ligation of DNA molecules (42).
Chen et al. identified three small molecules that inhibit human DNA Ligases by blocking their DBD. In particular, L82 inhibits LIG1, L67 inhibits LIG1 and LIG3, and L189 inhibits DNA ligases I, III and IV. Importantly, DNA Ligases inhibitors exert cytotoxic or cytostatic (L82) effects and sensitize cancer cells to DNA damaging agents (61). In addition, a recent report by Sallmyr et al. suggest that L67 preferentially targets mitochondrial LIG3 function in cancer cells, that in turn increased nuclear DNA damage and cell death, without any significant effects in nonmalignant cells (62).
Alt-NHEJ as therapeutic target in cancer: preclinical studies
In the following sections, we will describe the involvement of Alt-NHEJ in onset, progression and drug resistance of certain tumors.
Ovarian cancer
About half of epithelial ovarian cancers (EOCs) harbor deficit in HR repair that drives the genomic instability and addiction to PARP-directed DNA repair. Ceccaldi et al. (35) reported in EOCs patients an inverse correlation between HR activity and expression of PolQ. Moreover, HR-deficient tumor cells had higher steady state levels of PolQ, the DNA polymerase involved in Alt-NHEJ. In addition, the authors found that PolQ itself could inhibit HR, by blocking RAD51-mediated recombination. Importantly, knockdown of PolQ in HR-deficient EOCs reduced cell survival and genetic inactivation of FANCD2/BRCA2 and PolQ in mice results in embryonic lethality. Overall, these results confirmed a synthetic lethal relationship between the HR pathway and Alt-NHEJ repair in EOCs, characterizing PolQ as a new potential therapeutic target for HR-defective cancer.
Breast cancer
Alt-NHEJ is involved in breast cancer onset and acquisition of drug resistance. Indeed, Tobin et al demonstrated that estrogen receptor and progesterone receptor-positive (ER/PR+) MCF7 breast cancer cells have increased Alt-NHEJ pathway activity if compared to non-tumorigenic breast epithelial MCF10A cells. Moreover, the authors showed that up-regulation of core components LIG3 and PARP1 were observed in tamoxifen- and aromatase-resistant derivatives of MCF7 cells, resulting therefore in increased sensitivity to a combination of PARP and LIG3 inhibitors (63).
Neuroblastoma
Newman et al. showed that Alt-NHEJ is critical for neuroblastoma genomic instability and cell survival (64). Indeed, they demonstrated that an error prone DNA repair pathway was hyper-activated in neuroblastoma cells, which expressed low levels of mediators of C-NHEJ, LIG4 and Artemis (DCLRE1C) as compared to up-regulation observed for proteins required for Alt-NHEJ and PARP1, especially in MYCN overexpressing cell lines. Consistently, inhibition of LIG3 LIG1 by L67 and PARP1 by BYK204165 (BYK), led to DSB accumulation and cell death. These results were next confirmed by datasets analysis which revealed that higher levels of genes encoding Alt-NHEJ proteins were associated to poor overall survival. Moreover, the authors showed for the first time a crucial involvement of Alt-NHEJ in neuroblastoma initiation by mediating MYCN oncogenic activity in human neural crest stem cell differentiation (65).
Leukemia
There is experimental evidence showing that TK-activated leukemias are characterized by deep genomic instability.
Fan et al reported that in FLT3/ITD-expressing acute myeloid leukemia cell lines and bone marrow mononuclear cells from FLT3/ITD knock-in mice, Alt-NHEJ repair is hyper-activated resulting in a high frequency of DNA sequence aberrations. Core component of C-NHEJ Ku protein level was decreased while LIG3 was up-regulated in FLT3/ITD-expressing cells. Importantly, the authors showed that FLT3 signaling increased Alt-NHEJ repair activity and consistently, treatment with a FLT3 inhibitor (CEP-701) was able to decrease LIG3-mediated genomic instability in FLT3/ITD-expressing cells (66). Moreover, Hähnel et al. (67) demonstrated that oncogenic KRAS causes upregulation of components of the Alt-NHEJ pathway in T-cell acute lymphoblastic leukemia (T-ALL) and importantly the targeting of Alt-NHEJ pathway selectively sensitizes KRAS-mutant leukemic cells to cytotoxic agents such as cytarabine, daunorubicin or VP-16. Interestingly, Muvarak et al. showed that c-MYC contributed to genomic instability observed in TK-activated leukemias, by increasing the expression of LIG3 and PARP1, critical components of error prone Alt-NHEJ repair (68).
Sallmyr et al showed that also chronic myeloid leukemia (CML) BCR-ABL positive cells had an increased Alt-NHEJ repair activity. Indeed, key proteins of C-NHEJ pathway, Artemis and LIG4, were down-regulated, whereas LIG3 was up-regulated. Strikingly, knockdown of LIG3 led to increase of DNA damage, thus potentially impairing CML cell survival (69).
Multiple myeloma (MM)
MM is a hematologic malignancy characterized by abnormal proliferation of plasma cells harboring several chromosomal aberrations. Our group provided evidence that up-regulation of LIG3-mediated DNA repair plays a pivotal role in genomic instability and survival of MM cells (70). In particular, higher LIG3 mRNA expression in MM patients correlates with poor survival. Consistently knockdown of LIG3 impaired MM cells viability in vitro and in vivo, suggesting that malignant plasma cells are dependent on LIG3-driven repair. Importantly, we showed that LIG3 expression is under control of miR-22. Indeed, miR-22 replacement in MM cells down-regulated LIG3 protein and reduced Alt-NHEJ repair increasing DNA damage and cell death. Overall, these results indicate that myeloma cells are addicted to LIG3-driven Alt-NHEJ repair, which drive disease progression and drug resistance.
Moreover, results from targeted next generation sequencing studies in patients at all disease stages, demonstrated homologous recombination deficiency (HRD) in MM samples, which increased with progression of the disease, drug resistance acquisition and correlated with high-risk disease markers (71). Since Alt-NHEJ can compensate for HR defective repair, these data provide the rationale for evaluation of PARP inhibitors in MM patients, particularly in the relapsed setting. Indeed, preliminary results by our group show that Alt-NHEJ inhibitors are highly active against drug resistant MM cell lines and primary samples (Caracciolo et al., manuscript in preparation) further confirming the relevance of Alt-NHEJ for MM cells pathogenesis.
Conclusions and perspectives
Although dysregulation of the DDR represents a harmful mechanism by which cancer cells can acquire the mutator phenotype, at the same time it offers an intriguing area of investigation for therapeutic purpose. Indeed, cancer cells often have at least one deregulated DNA repair pathway and this deficiency been compensated by the activation of a second pathway, whose inhibition will specifically kill the malignant cells (synthetic lethality) with little effect on normal cells. Thus, this strategy provides a framework for the design of novel therapeutic approaches to selectively kill cancer cells, improving disease response and sparing patients from unnecessary side effects. However, the identification of biomarkers of DDR defects represents a challenge to overcome in the near future in order to explore other potential vulnerabilities in cancer biology.
In this field, the recent demonstration that cancer cells carrying defects in HR or NHEJ are more dependent upon a poorly defined Alt-NHEJ for DNA repair sheds new light on the mechanisms underpinning the efficacy of PARP-inhibitors in BRCA-defective ovarian and breast cancer and provides proof of concept for the design of innovative therapeutic strategies based on Alt-NHEJ inhibition. Indeed, in this review, we have described that elevated expression of core components, such as PARP-1, LIG3 and PolQ, occurs in several solid and haematologic malignancies with defects in the two major DSB repair pathways and could provide predictive biomarker for response to Alt-NHEJ inhibitors.
Although alteration of DNA repair genes could not account for all cases of genomic instability, especially in sporadic cancers, we reported evidence for a pivotal role of the Alt-NHEJ in large genomic rearrangements, in particular translocations, which underpins onset and progression of different tumors.
Since NGS technologies allows to characterize the status of individual DNA repair pathways in individual patients (72), the opportunity of combining Alt-NHEJ inhibitors with other treatment modalities (such as chemotherapy, immunotherapy or radiotherapy) in tumors with up-regulation of Alt-NHEJ, could be a promising strategy with potential application in the future.
In conclusion, this review underlines error-prone Alt-NHEJ repair pathway as an acheel heel for HR- and or NHEJ-defective cancer and therefore as a new opportunity for synthetic lethal approaches that can be exploited in the clinical scenario of precision oncology.
Acknowledgments
Funding: This work has been mainly supported by the Italian Association for Cancer Research (AIRC) with “Special Program for Molecular Clinical Oncology 5 per mille”, 2010/15 and its Extension Program” No. 9980, 2016/18 (PI: PT); and also by “Innovative Immunotherapeutic Treatments of Human Cancer” Multi Unit Regional No. 16695 (co-financed by AIRC and the CARICAL foundation). We thanks Dr. Ivana Criniti for her study coordination support and editorial assistance.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editor Masood A. Shammas for the series “Genomic Instability, Clonal Evolution and Oncogenesis” published in Precision Cancer Medicine. The article has undergone external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/pcm.2019.02.05). The series “Genomic Instability, Clonal Evolution and Oncogenesis” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability--an evolving hallmark of cancer. Nat Rev Mol Cell Biol 2010;11:220-8. [Crossref] [PubMed]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646-74. [Crossref] [PubMed]
- Dworaczek H, Xiao W. Xeroderma pigmentosum: a glimpse into nucleotide excision repair, genetic instability, and cancer. Crit Rev Oncog 2007;13:159-77. [Crossref] [PubMed]
- De' Angelis GL. Microsatellite instability in colorectal cancer. Acta Biomed 2018;89:97-101. [PubMed]
- Abida W, Cheng ML, Armenia J, et al. Analysis of the Prevalence of Microsatellite Instability in Prostate Cancer and Response to Immune Checkpoint Blockade. JAMA Oncol 2018; [Epub ahead of print]. [Crossref] [PubMed]
- Sheffer M, Bacolod MD, Zuk O, et al. Association of survival and disease progression with chromosomal instability: a genomic exploration of colorectal cancer. Proc Natl Acad Sci U S A 2009;106:7131-6. [Crossref] [PubMed]
- Fenech M. Chromosomal biomarkers of genomic instability relevant to cancer. Drug Discov Today 2002;7:1128-37. [Crossref] [PubMed]
- Sansregret L, Nepveu A. Gene signatures of genomic instability as prognostic tools for breast cancer. Future Oncol 2011;7:591-4. [Crossref] [PubMed]
- Fonseca R, Blood E, Rue M, et al. Clinical and biologic implications of recurrent genomic aberrations in myeloma. Blood 2003;101:4569-75. [Crossref] [PubMed]
- Murata S, Zhang C, Finch N, et al. Predictors and Modulators of Synthetic Lethality: An Update on PARP Inhibitors and Personalized Medicine. Biomed Res Int 2016;2016:2346585. [Crossref] [PubMed]
- Asaoka Y, Ijichi H, Koike K. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med 2015;373:1979. [Crossref] [PubMed]
- Jeggo PA, Pearl LH, Carr AM. DNA repair, genome stability and cancer: a historical perspective. Nat Rev Cancer 2016;16:35-42. [Crossref] [PubMed]
- Lindahl T, Barnes DE. Repair of endogenous DNA damage. Cold Spring Harb Symp Quant Biol 2000;65:127-33. [Crossref] [PubMed]
- Hoeijmakers JH. DNA damage, aging, and cancer. N Engl J Med 2009;361:1475-85. [Crossref] [PubMed]
- Pannunzio NR, Watanabe G, Lieber MR. Nonhomologous DNA end-joining for repair of DNA double-strand breaks. J Biol Chem 2018;293:10512-23. [Crossref] [PubMed]
- Burma S, Chen BP, Chen DJ. Role of non-homologous end joining (NHEJ) in maintaining genomic integrity. DNA Repair (Amst) 2006;5:1042-8. [Crossref] [PubMed]
- San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem 2008;77:229-57. [Crossref] [PubMed]
- Heyer WD, Ehmsen KT, Liu J. Regulation of homologous recombination in eukaryotes. Annu Rev Genet 2010;44:113-39. [Crossref] [PubMed]
- Wang H, Perrault AR, Takeda Y, et al. Biochemical evidence for Ku-independent backup pathways of NHEJ. Nucleic Acids Res 2003;31:5377-88. [Crossref] [PubMed]
- Terzoudi GI, Singh SK, Pantelias GE, et al. Premature chromosome condensation reveals DNA-PK independent pathways of chromosome break repair. Int J Oncol 2008;33:871-9. [PubMed]
- Chang HHY, Pannunzio NR, Adachi N, et al. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat Rev Mol Cell Biol 2017;18:495-506. [Crossref] [PubMed]
- Xiong X, Du Z, Wang Y, et al. 53BP1 promotes microhomology-mediated end-joining in G1-phase cells. Nucleic Acids Res 2015;43:1659-70. [Crossref] [PubMed]
- Xie A, Kwok A, Scully R. Role of mammalian Mre11 in classical and alternative nonhomologous end joining. Nat Struct Mol Biol 2009;16:814-8. [Crossref] [PubMed]
- Yang G, Liu C, Chen SH, et al. Super-resolution imaging identifies PARP1 and the Ku complex acting as DNA double-strand break sensors. Nucleic Acids Res 2018;46:3446-57. [Crossref] [PubMed]
- Anand R, Ranjha L, Cannavo E, et al. Phosphorylated CtIP Functions as a Co-factor of the MRE11-RAD50-NBS1 Endonuclease in DNA End Resection. Mol Cell 2016;64:940-50. [Crossref] [PubMed]
- Haince JF, McDonald D, Rodrigue A, et al. PARP1-dependent kinetics of recruitment of MRE11 and NBS1 proteins to multiple DNA damage sites. J Biol Chem 2008;283:1197-208. [Crossref] [PubMed]
- Kent T, Chandramouly G, McDevitt SM, et al. Mechanism of microhomology-mediated end-joining promoted by human DNA polymerase theta. Nat Struct Mol Biol 2015;22:230-7. [Crossref] [PubMed]
- Okano S, Lan L, Tomkinson AE, et al. Translocation of XRCC1 and DNA ligase IIIalpha from centrosomes to chromosomes in response to DNA damage in mitotic human cells. Nucleic Acids Res 2005;33:422-9. [Crossref] [PubMed]
- Della-Maria J, Zhou Y, Tsai MS, et al. Human Mre11/human Rad50/Nbs1 and DNA ligase IIIalpha/XRCC1 protein complexes act together in an alternative nonhomologous end joining pathway. J Biol Chem 2011;286:33845-53. [Crossref] [PubMed]
- Lu G, Duan J, Shu S, et al. Ligase I and ligase III mediate the DNA double-strand break ligation in alternative end-joining. Proc Natl Acad Sci U S A 2016;113:1256-60. [Crossref] [PubMed]
- Iliakis G, Murmann T, Soni A. Alternative end-joining repair pathways are the ultimate backup for abrogated classical non-homologous end-joining and homologous recombination repair: Implications for the formation of chromosome translocations. Mutat Res Genet Toxicol Environ Mutagen 2015;793:166-75. [Crossref] [PubMed]
- Paddock MN, Bauman AT, Higdon R, et al. Competition between PARP-1 and Ku70 control the decision between high-fidelity and mutagenic DNA repair. DNA Repair (Amst) 2011;10:338-43. [Crossref] [PubMed]
- Barton O, Naumann SC, Diemer-Biehs R, et al. Polo-like kinase 3 regulates CtIP during DNA double-strand break repair in G1. J Cell Biol 2014;206:877-94. [Crossref] [PubMed]
- Mao Z, Hine C, Tian X, et al. SIRT6 promotes DNA repair under stress by activating PARP1. Science 2011;332:1443-6. [Crossref] [PubMed]
- Ceccaldi R, Liu JC, Amunugama R, et al. Homologous-recombination-deficient tumours are dependent on Poltheta-mediated repair. Nature 2015;518:258-62. [Crossref] [PubMed]
- Mateos-Gomez PA, Kent T, Deng SK, et al. The helicase domain of Poltheta counteracts RPA to promote alt-NHEJ. Nat Struct Mol Biol 2017;24:1116-23. [Crossref] [PubMed]
- Dutta A, Eckelmann B, Adhikari S, et al. Microhomology-mediated end joining is activated in irradiated human cells due to phosphorylation-dependent formation of the XRCC1 repair complex. Nucleic Acids Res 2017;45:2585-99. [PubMed]
- Simsek D, Furda A, Gao Y, et al. Crucial role for DNA ligase III in mitochondria but not in Xrcc1-dependent repair. Nature 2011;471:245-8. [Crossref] [PubMed]
- Arana ME, Seki M, Wood RD, et al. Low-fidelity DNA synthesis by human DNA polymerase theta. Nucleic Acids Res 2008;36:3847-56. [Crossref] [PubMed]
- Wood RD, Doublie S. DNA polymerase theta (POLQ), double-strand break repair, and cancer. DNA Repair (Amst) 2016;44:22-32. [Crossref] [PubMed]
- Zhuang J, Jiang G, Willers H, et al. Exonuclease function of human Mre11 promotes deletional nonhomologous end joining. J Biol Chem 2009;284:30565-73. [Crossref] [PubMed]
- Taylor RM, Whitehouse CJ, Caldecott KW. The DNA ligase III zinc finger stimulates binding to DNA secondary structure and promotes end joining. Nucleic Acids Res 2000;28:3558-63. [Crossref] [PubMed]
- Simsek D, Brunet E, Wong SY, et al. DNA ligase III promotes alternative nonhomologous end-joining during chromosomal translocation formation. PLoS Genet 2011;7:e1002080. [Crossref] [PubMed]
- Panchakshari RA, Zhang X, Kumar V, et al. DNA double-strand break response factors influence end-joining features of IgH class switch and general translocation junctions. Proc Natl Acad Sci U S A 2018;115:762-7. [Crossref] [PubMed]
- Kotnis A, Du L, Liu C, et al. Non-homologous end joining in class switch recombination: the beginning of the end. Philos Trans R Soc Lond B Biol Sci 2009;364:653-65. [Crossref] [PubMed]
- Yan CT, Boboila C, Souza EK, et al. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature 2007;449:478-82. [Crossref] [PubMed]
- Zhu C, Mills KD, Ferguson DO, et al. Unrepaired DNA breaks in p53-deficient cells lead to oncogenic gene amplification subsequent to translocations. Cell 2002;109:811-21. [Crossref] [PubMed]
- Zhu C, Bogue MA, Lim DS, et al. Ku86-deficient mice exhibit severe combined immunodeficiency and defective processing of V(D)J recombination intermediates. Cell 1996;86:379-89. [Crossref] [PubMed]
- Taccioli GE, Amatucci AG, Beamish HJ, et al. Targeted disruption of the catalytic subunit of the DNA-PK gene in mice confers severe combined immunodeficiency and radiosensitivity. Immunity 1998;9:355-66. [Crossref] [PubMed]
- Wray J, Williamson EA, Singh SB, et al. PARP1 is required for chromosomal translocations. Blood 2013;121:4359-65. [Crossref] [PubMed]
- Krishnakumar R, Kraus WL. The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets. Mol Cell 2010;39:8-24. [Crossref] [PubMed]
- Morales J, Li L, Fattah FJ, et al. Review of poly (ADP-ribose) polymerase (PARP) mechanisms of action and rationale for targeting in cancer and other diseases. Crit Rev Eukaryot Gene Expr 2014;24:15-28. [Crossref] [PubMed]
- O'Sullivan CC, Moon DH, Kohn EC, et al. Beyond Breast and Ovarian Cancers: PARP Inhibitors for BRCA Mutation-Associated and BRCA-Like Solid Tumors. Front Oncol 2014;4:42. [PubMed]
- Farmer H, McCabe N, Lord CJ, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 2005;434:917-21. [Crossref] [PubMed]
- Bryant HE, Schultz N, Thomas HD, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 2005;434:913-7. [Crossref] [PubMed]
- Scott CL, Swisher EM, Kaufmann SH. Poly (ADP-ribose) polymerase inhibitors: recent advances and future development. J Clin Oncol 2015;33:1397-406. [Crossref] [PubMed]
- Gottipati P, Vischioni B, Schultz N, et al. Poly(ADP-ribose) polymerase is hyperactivated in homologous recombination-defective cells. Cancer Res 2010;70:5389-98. [Crossref] [PubMed]
- Lakshmipathy U, Campbell C. The human DNA ligase III gene encodes nuclear and mitochondrial proteins. Mol Cell Biol 1999;19:3869-76. [Crossref] [PubMed]
- Wang H, Rosidi B, Perrault R, et al. DNA ligase III as a candidate component of backup pathways of nonhomologous end joining. Cancer Res 2005;65:4020-30. [Crossref] [PubMed]
- Akbari M, Keijzers G, Maynard S, et al. Overexpression of DNA ligase III in mitochondria protects cells against oxidative stress and improves mitochondrial DNA base excision repair. DNA Repair (Amst) 2014;16:44-53. [Crossref] [PubMed]
- Chen X, Zhong S, Zhu X, et al. Rational design of human DNA ligase inhibitors that target cellular DNA replication and repair. Cancer Res 2008;68:3169-77. [Crossref] [PubMed]
- Sallmyr A, Matsumoto Y, Roginskaya V, et al. Inhibiting Mitochondrial DNA Ligase IIIalpha Activates Caspase 1-Dependent Apoptosis in Cancer Cells. Cancer Res 2016;76:5431-41. [Crossref] [PubMed]
- Tobin LA, Robert C, Nagaria P, et al. Targeting abnormal DNA repair in therapy-resistant breast cancers. Mol Cancer Res 2012;10:96-107. [Crossref] [PubMed]
- Newman EA, Lu F, Bashllari D, et al. Alternative NHEJ Pathway Components Are Therapeutic Targets in High-Risk Neuroblastoma. Mol Cancer Res 2015;13:470-82. [Crossref] [PubMed]
- Newman EA, Chukkapalli S, Bashllari D, et al. Alternative NHEJ pathway proteins as components of MYCN oncogenic activity in human neural crest stem cell differentiation: implications for neuroblastoma initiation. Cell Death Dis 2017;8:3208. [Crossref] [PubMed]
- Fan J, Li L, Small D, et al. Cells expressing FLT3/ITD mutations exhibit elevated repair errors generated through alternative NHEJ pathways: implications for genomic instability and therapy. Blood 2010;116:5298-305. [Crossref] [PubMed]
- Hähnel PS, Enders B, Sasca D, et al. Targeting components of the alternative NHEJ pathway sensitizes KRAS mutant leukemic cells to chemotherapy. Blood 2014;123:2355-66. [Crossref] [PubMed]
- Muvarak N, Kelley S, Robert C, et al. c-MYC Generates Repair Errors via Increased Transcription of Alternative-NHEJ Factors, LIG3 and PARP1, in Tyrosine Kinase-Activated Leukemias. Mol Cancer Res 2015;13:699-712. [Crossref] [PubMed]
- Sallmyr A, Tomkinson AE, Rassool FV. Up-regulation of WRN and DNA ligase IIIalpha in chronic myeloid leukemia: consequences for the repair of DNA double-strand breaks. Blood 2008;112:1413-23. [Crossref] [PubMed]
- Caracciolo D, Di Martino MT, Amodio N, et al. miR-22 suppresses DNA ligase III addiction in multiple myeloma. Leukemia 2019;33:487-98. [Crossref] [PubMed]
- Pawlyn C, Loehr A, Ashby C, et al. Loss of heterozygosity as a marker of homologous repair deficiency in multiple myeloma: a role for PARP inhibition? Leukemia 2018;32:1561-6. [Crossref] [PubMed]
- Meier B, Gartner A. Having a direct look: analysis of DNA damage and repair mechanisms by next generation sequencing. Exp Cell Res 2014;329:35-41. [Crossref] [PubMed]
Cite this article as: Caracciolo D, Montesano M, Tagliaferri P, Tassone P. Alternative non-homologous end joining repair: a master regulator of genomic instability in cancer. Precis Cancer Med 2019;2:8.

