
The structure of mTOR complexes at a glance
Rapamycin initially isolated from Rapa Nui island soil sample, has anti-fungal, immunosuppressive and antitumor properties (1,2). A rapamycin targeting protein, target of rapamycin (TOR), was first identified in yeast in 1991 (3). Later, the mechanistic (formerly “mammalian”) TOR (mTOR) in mammals was identified by biochemical studies (4,5). Rapamycin, in complex with the endogenous protein FKBP12, inhibits mTOR complex 1 (mTORC1) kinase activity through binding the FKBP-rapamycin binding domain (FRB) of mTOR (4). After two decades, studies from many groups have revealed that mTOR is a serine/threonine protein kinase belonging to phosphatidylinositol 3-kinase (PI3K)-related kinase (PIKK) family and is the central regulator of cell growth and metabolism (6).
In mammals, mTOR exists in two complexes: rapamycin-sensitive mTOR complex 1 (mTORC1) and rapamycin-insensitive mTOR complex 2 (mTORC2). The two complexes share mTOR, mLST8 (mammalian homolog of protein Lethal with Sec13 protein 8) core subunits and an additional non-core subunit Deptor (DEP-domain-containing mTOR-interacting protein), and differ in Raptor (regulatory-associated protein of mTOR) and Rictor (rapamycin-insensitive companion of mTOR) (7,8). Besides, mTORC1 complex contains an inhibitory regulator protein PRAS40 (proline-rich AKT substrate of 40 KD). mTORC2 complex contains an additional core subunit: mSin1, which is indispensable for the complex integrity and a non-core subunit Protor-1 (protein observed with Rictor-1) (9). There are two TOR genes in yeast, TOR1 and TOR2. Both TOR1 and TOR2 can form the catalytic core of TORC1. However, only TOR2 can form the catalytic core of TORC2 (10,11).
In response to environmental nutrients and energy signals, mTORC1 inhibits cell growth through phosphorylation of downstream targets, primarily the 70S ribosomal protein S6 kinase (S6K1) and the eukaryotic initiation factor 4E binding protein (4E-BP1) (12,13). Dysregulation of mTORC1 is often associated with the development of cancer, diabetes and neurological disease (10). In contrast to mTORC1, mTORC2 controls cell proliferation, survival and cytoskeletal remodeling through phosphorylation of AGC and PKC kinase family members, such as AKT (8).
The functions of mTOR complexes have been extensively studied in the past two decades. However, the structural information of mTORC1 or mTORC2 was limited to low resolution because of technical challenges. It’s very difficult to obtain highly purified and homogeneous protein samples given the fact that mTORC1 and mTORC2 form complexes with molecular weight over 1 MD (1,000,000 Dalton) (14). Consistent with their high molecular weights, both complexes are not ideal for structural studies using X-ray crystallography. We and other groups took advantage of the recent breakthrough in both sample preparation using mammalian and insect cell expression and cryo-electron microscopy (cryo-EM) technology to make progress in improving the resolution of mTORC1 and mTORC2 structures (Figure 1).

The first cryo-EM structure of mTORC1 was determined at ~26 Å resolution, presenting limited structural information (15). In 2013, Pavletich lab reported the crystal structure of mTOR C-terminal fragment containing the FAT and kinase domain in complex with mLST8 (16). In three recent studies, the resolution of cryo-EM structures of human mTORC1 complex were improved to 5.9 Å, 4.4 Å, and 3.2 Å, respectively (18,20,21). The yeast Tor-Lst8 structure was determined at 6 Å resolution. By inserting Red Fluorescent Proteins (RFP) (19), they experimentally determined the topology of mTOR, which is consistent with the mTORC1 structure at 4.4 Å (20), but distinct from the previously reported mTORC1 structure at 5.9 Å (18). Finally, Pavletich lab presented an elaborate work of the mTORC1 structure as well as the activation and inhibition mechanism of mTORC1 activity by Rheb (Ras homolog enriched in brain) and PRAS40 respectively (21).
As shown in Figure 2, mTOR consists of an N-terminal HEAT (Huntington, EF3A, ATM, TOR) repeat (N-HEAT), a middle HEAT (M-HEAT), an FAT (Frap, ATM, TRRAP), an FRB (FKBP12-Rapamycin binding domain) and a C-terminal kinase domain. Raptor is responsible for substrate recruitment and contains an N-terminal caspase-like domain, HEAT repeats and C-terminal WD40 repeats. mLST8 is primarily composed of WD40 repeats and is an indispensable subunit for mTORC1/2 complexes (20). Rictor has three continuous ARM/HEAT helical repeat (HR) clusters (HR1 to HR3), followed by a large unstructured region. mSin1 is composed of an N-terminal region, a CRIM (conserved region in the middle), an RBD (Ras binding domain), and a PH (pleckstrin homology domain) (22) (Figure 2).

The mTORC1 adopts a hollow rhombohedral shape and forms a symmetric dimer of heterotrimer (mTOR-Raptor-mLST8) mediated by two mTOR monomers and is stabilized by the Raptor subunit that binds across both monomers. The N-HEAT of mTOR adopts a spiral (~1.3-turn) right-hand superhelix comprising 16 HEAT repeats. The M-HEAT bridges the N-HEAT and the FAT/kinase module. The C-terminal region of mTOR forms a compact core domain, in which the α-solenoid (FAT domain) as a “C”-shape wraps around the kinase domain from which the FRB domain protrudes out. Although Raptor has a conserved N-terminal caspase-like domain, it shows no detectable caspase activity. The pseudo catalytic pocket of the caspase-like domain faces toward the catalytic cavity of the kinase domain of mTOR, supporting its function in mediating substrate recruitment for phosphorylation (20). The mTOR kinase domain shows the characteristic two-lobe structure of PI3K kinase, which consists of an N-terminal lobe (N lobe), a larger C-terminal lobe (C lobe) and a cleft between the two lobes that binds to ATP (24). Comparing to the PI3K kinase domain, mTOR kinase domain contains an FRB domain inserted within the N lobe and a 40-residue-insertion in the C lobe that contributes to the association of mLST8. mLST8 is located on the distal convex along the short axis and binds the C lobe to stabilize the kinase active pocket (16) (Figure 3).
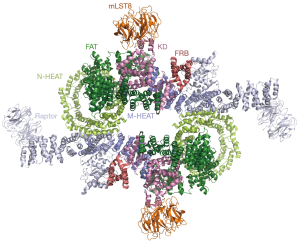
RHEB is a small GTPase protein and plays an important role in activating mTORC1 activity in vivo (25). The activation mechanism is beautifully revealed by the cryo-EM structure of RHEB-mTORC1 complex (21). As shown in Figure 4, RHEB binds to the N-HEAT, M-HEAT, and FAT domains of mTOR. Upon RHEB binding, the N-HEAT solenoid rotates and moves toward the M-HEAT, which facilitates interactions between the N-terminal portions of the N-HEAT and the FAT domain. Thus, the middle portion of FAT domain gets twisted and dragged by the N-HEAT solenoid. This conformational change leads to a less compact interaction between the FAT and N-lobe of the kinase domain and a narrower catalytic cleft between the N- and C-lobes. Therefore, RHEB is likely to activate mTORC1 by allosterically realigning the active site residues for catalysis. Interestingly, cancer-associated mTOR mutants located at the major intra-FAT hinge, the FAT-N lobe packing transition, and the N-lobe anchor in a pocket between the C-lobe and FAT can hyperactive mTOR activity. These mutants may mimic RHEB activation on mTOR activity (Figure 4).
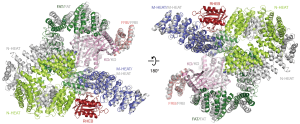
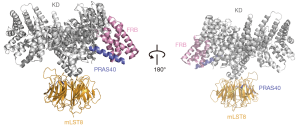
The crystal structure of S6K1-FRB suggests a second substrate binding site on the surface of FRB except Raptor-TOS (Tor signaling sequence) interface for the binding between mTORC1 and the substrates (21). PRAS40 functions as an inhibitor of mTORC1 activity (26,27). The co-crystal structure of PRAS40 (residues 173–256) with mTORΔN-mLST8 shows that PRAS40 binds mTORC1 through an amphipathic α-helix (residues 212–232) bound to FRB domain and a β-strand (residues 188–196) bound to mLST8 WD40 domain (Figure 5). S6K1 binds to the same site on the FRB with a binding affinity lower than that of PRAS40. The mutations of FRB-interacting residues of PRAS40 reduce the inhibition of mTORC1 kinase activity against 4E-BP1. Thus, PRAS40 leads to the inhibition through competitively blocking the substrates recruitment (21).
Negative stain EM (~26 Å) and cryo-EM (7.9 Å) structures of S. cerevisiae TORC2 (scTORC2) revealed an overall fold and subunit architecture of the complex (17,28). Recently, the cryo-EM structures of human mTORC2 were determined at 4.9 Å and 7.4 Å resolution, respectively (22,23).
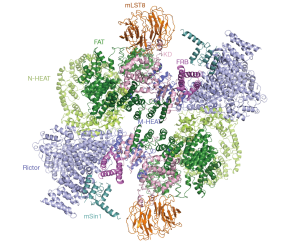
The mTORC2 forms a symmetric dimer of heterotetramer (mTOR-Rictor-mLST8-mSin1) and adopts a hollow rhombohedral fold with an mTOR-dimer served as a central scaffold. The inner hole of mTORC2 is as narrow as 11 Å, whereas that of mTORC1 is about 23 Å, showing a more compact fold than mTORC1. The structural model was built based on a combination of biochemistry, XL-MS and the cryo-EM map (22). As in mTORC1, mLST8 stably binds to the kinase domain of mTOR. The N-terminal helical repeat cluster of Rictor binds to mTOR through multiple contacts. The N-terminus of mSin1 is located close to the FRB domain and the catalytic cavity of mTOR, in agreement with its essential role in mediating the complex integrity (Figure 6). Structural comparison of mTORC2 and FRB-rapamycin-FKBP12 (PDB: 1FAP) (29) clearly shows that the α-helices of mSin1 N-terminus generates steric hindrance and would inhibit interaction between FKBP12-rapamycin and mTOR. The structural and biochemical analyses nicely explain the mechanism for rapamycin-insensitivity of mTORC2 (22).
Although the recent progresses were achieved on the structure of mTOR complexes, there are still some key questions need to be addressed. Such as the activation of mTORC2 signaling by PI3K. PtdIns(3,4,5)P3 is generated upon insulin or growth stimulation by activated PI3K. It was reported that mSin1-PH domain interacts with the mTOR kinase domain to suppress mTORC2 activity, and PtdIns(3,4,5)P3 binds the mSin1-PH domain to release its inhibition on the mTOR kinase domain, leading to mTORC2 activation (30). While an in vitro kinase assay using purified mTORC2 complexes containing various C-terminal truncations of mSin1 shows no significant difference in catalytic activity for the complex formed by full-length mSin1 or truncated mSin1 lacking PH domain. One possible explanation is that mTORC2 protein complex was obtained from 293F suspension cells cultured in serum-free medium. Under such condition, mTORC2 protein might adopt an active conformation in which the PH-mediated autoinhibition does not exist for unknown reason (22). So structural study of mTORC2 complex with further high-resolution and full-length mSin1 in an inhibitory conformation is needed to illustrate this question.
In the past decades, many inhibitors have been discovered or developed targeting mTOR complexes for research and/or clinical applications. The representative inhibitors include the allosteric inhibitors (Rapamycin and its derivatives) and ATP-competitive inhibitors (Torin, AZD8055 and CC-223, etc.) (31).
As shown above, rapamycin specifically inhibits mTORC1 enzymatic activity. Rapamycin can’t completely impair the activity of mTORC1 compared with ATP-competitive inhibitors (15). Rapamycin derivatives can be designed based on the structure of mTOR. Everolimus (RAD-001, Novartis), a rapamycin derivative, showed unsatisfactory efficacy in the global phase III randomized EVOLVE-1 (EVerOlimus for LiVer cancer Evaluation-1) trial, suggesting a potential mechanism for drug resistance against mTORC1 inhibitors in HCC (hepatocellular carcinoma). One reason for rapamycin derivatives failure in clinical trial is that they are substrate-selective mTORC1 inhibitors. They can inhibit S6K1 phosphorylation, but only show partially block on 4E-BP1 phosphorylation and cap-dependent translation (32). The crystal structure of FRB-S6K1 peptide shows that FRB, as the second binding site for S6K1, counts largely on the activity of mTORC1 phosphorylating S6K1. There is no direct structural evidence for FRB-4E-BP1, but the mutation of FRB-interacting residues of inhibitory protein PRAS40 reduced inhibition of mTORC1 phosphorylating 4E-BP1 by a factor of approximately 50 (21). The result shows that FRB might also interact with 4E-BP1. Therefore, testing 4E-BP1 binding with FRB or an FRB-4E-BP1 structure may provide the molecular insights into how mTORC1 and 4E-BP1 interact.
Phosphorylation of S6K1 by mTORC1 not only results in protein translation but also creates a negative feedback loop whereby the phosphorylated S6K1 decreases PI3K signaling and leads to mTORC1 inhibition (33). Thus, mTORC1 inhibition by rapamycin and its derivatives disrupt S6K1-mediated feedback inhibition of PI3K signaling and lead to increased PKB/AKT phosphorylation (27). Since mTORC2 is an upstream activator of the AGC kinase Akt and functions downstream of PI3K signaling. Therefore, dual mTORC1 and mTORC2 inhibition may be an attractive pharmacologic target with therapeutic potential in advanced HCC treatment. It is reported that dual targeting of mTORC1/C2 complexes enhances histone deacetylase inhibitor-mediated anti-tumor efficacy in primary HCC cancer in vitro and in vivo (34).
The development of ATP-competitive small molecule inhibitors targeting mTOR kinase, which inhibit both mTORC1 and mTORC2 are being developed as candidate anticancer agents, such as Torin (35). AZD8055 is the first drug that inhibits both types of mTOR complexes and is expected to be more effective than prior mTOR inhibitors, which is used for phase I in malignant gliomas (36). Another candidate, CC-223, was modified by structure-activity relationship and used for phase I expansion trial in advanced solid tumors (37,38).
An mTORC2 specific inhibitor could be useful to inhibit AKT activation and possesses potential clinical value (39). To avoid inhibition of mTORC1, such inhibitors should not be designed to target the mTOR kinase domain, a shared module within mTORC1 and mTORC2. According to the mTORC2 structure, the mTORC2 specific inhibitor could be designed to disrupt protein-protein interactions essential for mTORC2 integrity or the substrate-enzyme binding interface. Thus, the higher-resolution structure of mTORC2 and mTORC2-AKT complex are needed for structure-guided drug design.
Given the important function of mTOR complexes in regulating cell proliferation, there are many drugs targeting mTOR complexes were developed in the past two decades. The use of single mTOR inhibitor may cause negative feedback loops of PI3K. Thus, the combinational uses of mTOR kinase inhibitors and PI3K inhibitors for cancers are proved to be more potent in clinical trials.
Acknowledgments
We apologize to those authors whose works were not included here due to limited space.
Funding: This work was supported by grants from the Ministry of Science and Technology of China (2016YFA0500700), the National Natural Science Foundation of China (31770781, U1432242, 31425008, 91419301), the National Program for support of Top-Notch Young Professionals (Y Xu), and the Strategic Priority Research Program of the Chinese Academy of Sciences (grant no. XDB08000000).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/pcm.2018.07.02). YX serves as an unpaid editorial board member of Precision Cancer Medicine from Apr 2018 to Mar 2023. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Eng CP, Sehgal SN, Vezina C. Activity of rapamycin (AY-22,989) against transplanted tumors. J Antibiot (Tokyo) 1984;37:1231-7. [Crossref] [PubMed]
- Martel RR, Klicius J, Galet S. Inhibition of the immune response by rapamycin, a new antifungal antibiotic. Can J Physiol Pharmacol 1977;55:48-51. [Crossref] [PubMed]
- Heitman J, Movva NR, Hall MN. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science 1991;253:905-9. [Crossref] [PubMed]
- Sabatini DM, Erdjument-Bromage H, Lui M, et al. RAFT1: a mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell 1994;78:35-43. [Crossref] [PubMed]
- Brown EJ, Albers MW, Shin TB, et al. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature 1994;369:756-8. [Crossref] [PubMed]
- Saxton RA, Sabatini DM. mTOR Signaling in Growth, Metabolism, and Disease. Cell 2017;168:960-76. [Crossref] [PubMed]
- Kim DH, Sarbassov DD, Ali SM, et al. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 2002;110:163-75. [Crossref] [PubMed]
- Jacinto E, Loewith R, Schmidt A, et al. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat Cell Biol 2004;6:1122-8. [Crossref] [PubMed]
- Frias MA, Thoreen CC, Jaffe JD, et al. mSin1 is necessary for Akt/PKB phosphorylation, and its isoforms define three distinct mTORC2s. Curr Biol 2006;16:1865-70. [Crossref] [PubMed]
- Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell 2006;124:471-84. [Crossref] [PubMed]
- Loewith R, Jacinto E, Wullschleger S, et al. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol Cell 2002;10:457-68. [Crossref] [PubMed]
- Nojima H, Tokunaga C, Eguchi S, et al. The mammalian target of rapamycin (mTOR) partner, raptor, binds the mTOR substrates p70 S6 kinase and 4E-BP1 through their TOR signaling (TOS) motif. J Biol Chem 2003;278:15461-4. [Crossref] [PubMed]
- Schalm SS, Fingar DC, Sabatini DM, et al. TOS motif-mediated raptor binding regulates 4E-BP1 multisite phosphorylation and function. Curr Biol 2003;13:797-806. [Crossref] [PubMed]
- Wullschleger S, Loewith R, Oppliger W, et al. Molecular organization of target of rapamycin complex 2. J Biol Chem 2005;280:30697-704. [Crossref] [PubMed]
- Yip CK, Murata K, Walz T, et al. Structure of the human mTOR complex I and its implications for rapamycin inhibition. Mol Cell 2010;38:768-74. [Crossref] [PubMed]
- Yang H, Rudge DG, Koos JD, et al. mTOR kinase structure, mechanism and regulation. Nature 2013;497:217-23. [Crossref] [PubMed]
- Gaubitz C, Oliveira TM, Prouteau M, et al. Molecular Basis of the Rapamycin Insensitivity of Target Of Rapamycin Complex 2. Mol Cell 2015;58:977-88. [Crossref] [PubMed]
- Aylett CH, Sauer E, Imseng S, et al. Architecture of human mTOR complex 1. Science 2016;351:48-52. [Crossref] [PubMed]
- Baretić D, Berndt A, Ohashi Y, et al. Tor forms a dimer through an N-terminal helical solenoid with a complex topology. Nat Commun 2016;7:11016. [Crossref] [PubMed]
- Yang H, Wang J, Liu M, et al. 4.4 A Resolution Cryo-EM structure of human mTOR Complex 1. Protein Cell 2016;7:878-87. [Crossref] [PubMed]
- Yang H, Jiang X, Li B, et al. Mechanisms of mTORC1 activation by RHEB and inhibition by PRAS40. Nature 2017;552:368-73. [Crossref] [PubMed]
- Chen X, Liu M, Tian Y, et al. Cryo-EM structure of human mTOR complex 2. Cell Res 2018;28:518-28. [Crossref] [PubMed]
- Stuttfeld E, Aylett CH, Imseng S, et al. Architecture of the human mTORC2 core complex. Elife 2018;7. [PubMed]
- Walker EH, Perisic O, Ried C, et al. Structural insights into phosphoinositide 3-kinase catalysis and signalling. Nature 1999;402:313-20. [Crossref] [PubMed]
- Long X, Lin Y, Ortiz-Vega S, et al. Rheb binds and regulates the mTOR kinase. Curr Biol 2005;15:702-13. [Crossref] [PubMed]
- Wang L, Harris TE, Roth RA, et al. PRAS40 regulates mTORC1 kinase activity by functioning as a direct inhibitor of substrate binding. J Biol Chem 2007;282:20036-44. [Crossref] [PubMed]
- Wan X, Harkavy B, Shen N, et al. Rapamycin induces feedback activation of Akt signaling through an IGF-1R-dependent mechanism. Oncogene 2007;26:1932-40. [Crossref] [PubMed]
- Karuppasamy M, Kusmider B, Oliveira TM, et al. Cryo-EM structure of Saccharomyces cerevisiae target of rapamycin complex 2. Nat Commun 2017;8:1729. [Crossref] [PubMed]
- Choi J, Chen J, Schreiber SL, et al. Structure of the FKBP12-rapamycin complex interacting with the binding domain of human FRAP. Science 1996;273:239-42. [Crossref] [PubMed]
- Liu P, Gan W, Chin YR, et al. PtdIns(3,4,5)P3-Dependent Activation of the mTORC2 Kinase Complex. Cancer Discov 2015;5:1194-209. [Crossref] [PubMed]
- Zheng Y, Jiang Y. mTOR Inhibitors at a Glance. Mol Cell Pharmacol 2015;7:15-20. [PubMed]
- Choo AY, Yoon SO, Kim SG, et al. Rapamycin differentially inhibits S6Ks and 4E-BP1 to mediate cell-type-specific repression of mRNA translation. Proc Natl Acad Sci U S A 2008;105:17414-9. [Crossref] [PubMed]
- Manning BD. Balancing Akt with S6K: implications for both metabolic diseases and tumorigenesis. J Cell Biol 2004;167:399-403. [Crossref] [PubMed]
- Shao H, Gao C, Tang H, et al. Dual targeting of mTORC1/C2 complexes enhances histone deacetylase inhibitor-mediated anti-tumor efficacy in primary HCC cancer in vitro and in vivo. J Hepatol 2012;56:176-83. [Crossref] [PubMed]
- Thoreen CC, Kang SA, Chang JW, et al. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem 2009;284:8023-32. [Crossref] [PubMed]
- Luchman HA, Stechishin OD, Nguyen SA, et al. Dual mTORC1/2 blockade inhibits glioblastoma brain tumor initiating cells in vitro and in vivo and synergizes with temozolomide to increase orthotopic xenograft survival. Clin Cancer Res 2014;20:5756-67. [Crossref] [PubMed]
- Mortensen DS, Fultz KE, Xu S, et al. CC-223, a Potent and Selective Inhibitor of mTOR Kinase: In Vitro and In Vivo Characterization. Mol Cancer Ther 2015;14:1295-305. [Crossref] [PubMed]
- Bendell JC, Kelley RK, Shih KC, et al. A phase I dose-escalation study to assess safety, tolerability, pharmacokinetics, and preliminary efficacy of the dual mTORC1/mTORC2 kinase inhibitor CC-223 in patients with advanced solid tumors or multiple myeloma. Cancer 2015;121:3481-90. [Crossref] [PubMed]
- Sparks CA, Guertin DA. Targeting mTOR: prospects for mTOR complex 2 inhibitors in cancer therapy. Oncogene 2010;29:3733-44. [Crossref] [PubMed]
Cite this article as: Yang H, Chen X, Liu M, Xu Y. The structure of mTOR complexes at a glance. Precis Cancer Med 2018;1:7.

